Drop-seq Example
Last updated: 2018-08-29
workflowr checks: (Click a bullet for more information)-
✖ R Markdown file: uncommitted changes
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can runwflow_publishto commit the R Markdown file and build the HTML. -
✔ Environment: empty
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
-
✔ Seed:
set.seed(20180618)The command
set.seed(20180618)was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible. -
✔ Session information: recorded
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
-
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.✔ Repository version: f6c8902
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can usewflow_publishorwflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.Ignored files: Ignored: .Rhistory Ignored: .Rproj.user/ Ignored: R/.Rhistory Ignored: analysis/.Rhistory Ignored: analysis/pipeline/.Rhistory Untracked files: Untracked: ..gif Untracked: .DS_Store Untracked: R/.DS_Store Untracked: R/myheatmap.R Untracked: analysis/.DS_Store Untracked: analysis/Dropseqpreprocessing_cache/ Untracked: analysis/SLSL_marker_based_logcpm.pdf Untracked: analysis/biomarkers.R Untracked: analysis/cellref.pdf Untracked: analysis/consistency_check.R Untracked: analysis/dropseq_sc3_result.Rdata Untracked: analysis/dropseq_slsl1.Rdata Untracked: analysis/example_dropseq_cache/ Untracked: analysis/example_smartseq.Rmd Untracked: analysis/figure/ Untracked: analysis/marker-based.pdf Untracked: analysis/normalization_test.R Untracked: analysis/pbmcheat.pdf Untracked: analysis/pbmcref.pdf Untracked: analysis/pipeline/0_dropseq/ Untracked: analysis/pipeline/1_10X/ Untracked: analysis/pipeline/2_zeisel/ Untracked: analysis/pipeline/3_smallsets/ Untracked: analysis/writeup/cite.log Untracked: analysis/writeup/paper.aux Untracked: analysis/writeup/paper.bbl Untracked: analysis/writeup/paper.blg Untracked: analysis/writeup/paper.log Untracked: analysis/writeup/paper.out Untracked: analysis/writeup/paper.synctex.gz Untracked: analysis/writeup/paper.tex Untracked: analysis/writeup/writeup.aux Untracked: analysis/writeup/writeup.bbl Untracked: analysis/writeup/writeup.blg Untracked: analysis/writeup/writeup.dvi Untracked: analysis/writeup/writeup.log Untracked: analysis/writeup/writeup.out Untracked: analysis/writeup/writeup.synctex.gz Untracked: analysis/writeup/writeup.tex Untracked: analysis/writeup/writeup2.aux Untracked: analysis/writeup/writeup2.bbl Untracked: analysis/writeup/writeup2.blg Untracked: analysis/writeup/writeup2.log Untracked: analysis/writeup/writeup2.out Untracked: analysis/writeup/writeup2.pdf Untracked: analysis/writeup/writeup2.synctex.gz Untracked: analysis/writeup/writeup2.tex Untracked: analysis/writeup/writeup3.aux Untracked: analysis/writeup/writeup3.log Untracked: analysis/writeup/writeup3.out Untracked: analysis/writeup/writeup3.synctex.gz Untracked: analysis/writeup/writeup3.tex Untracked: data/unnecessary_in_building/ Untracked: normalized_out_1.Rdata Untracked: not_normalized_out_1.Rdata Untracked: src/.gitignore Untracked: tutorial2.Rmd Unstaged changes: Modified: NAMESPACE Modified: R/RcppExports.R Deleted: analysis/10Xpreprocessing.Rmd Deleted: analysis/Dropseqpreprocessing.Rmd Modified: analysis/_site.yml Modified: analysis/about.Rmd Modified: analysis/example_10x.Rmd Modified: analysis/example_dropseq.Rmd Modified: analysis/license.Rmd Modified: analysis/pipeline/.DS_Store Modified: analysis/writeup/.DS_Store Modified: data/.DS_Store
Expand here to see past versions:
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | f6c8902 | tk382 | 2018-08-29 | Build site. |
Read data.
library(data.table)
tmp = setDF(fread('data/unnecessary_in_building/GSM1626794_P14Retina_2.digital_expression.txt'))
orig = tmp[,-1]; rownames(orig) = tmp[,1]; rm(tmp)
genenames = sapply(strsplit(rownames(orig), ":"), function(x) x[3])
gc(verbose=FALSE); used (Mb) gc trigger (Mb) limit (Mb) max used (Mb)
Ncells 2792253 149.2 4257502 227.4 NA 4257502 227.4
Vcells 106691790 814.0 155381158 1185.5 16384 107648931 821.3Quality Control and Cell Filter
nGene = colSums(orig > 0)
hist(nGene)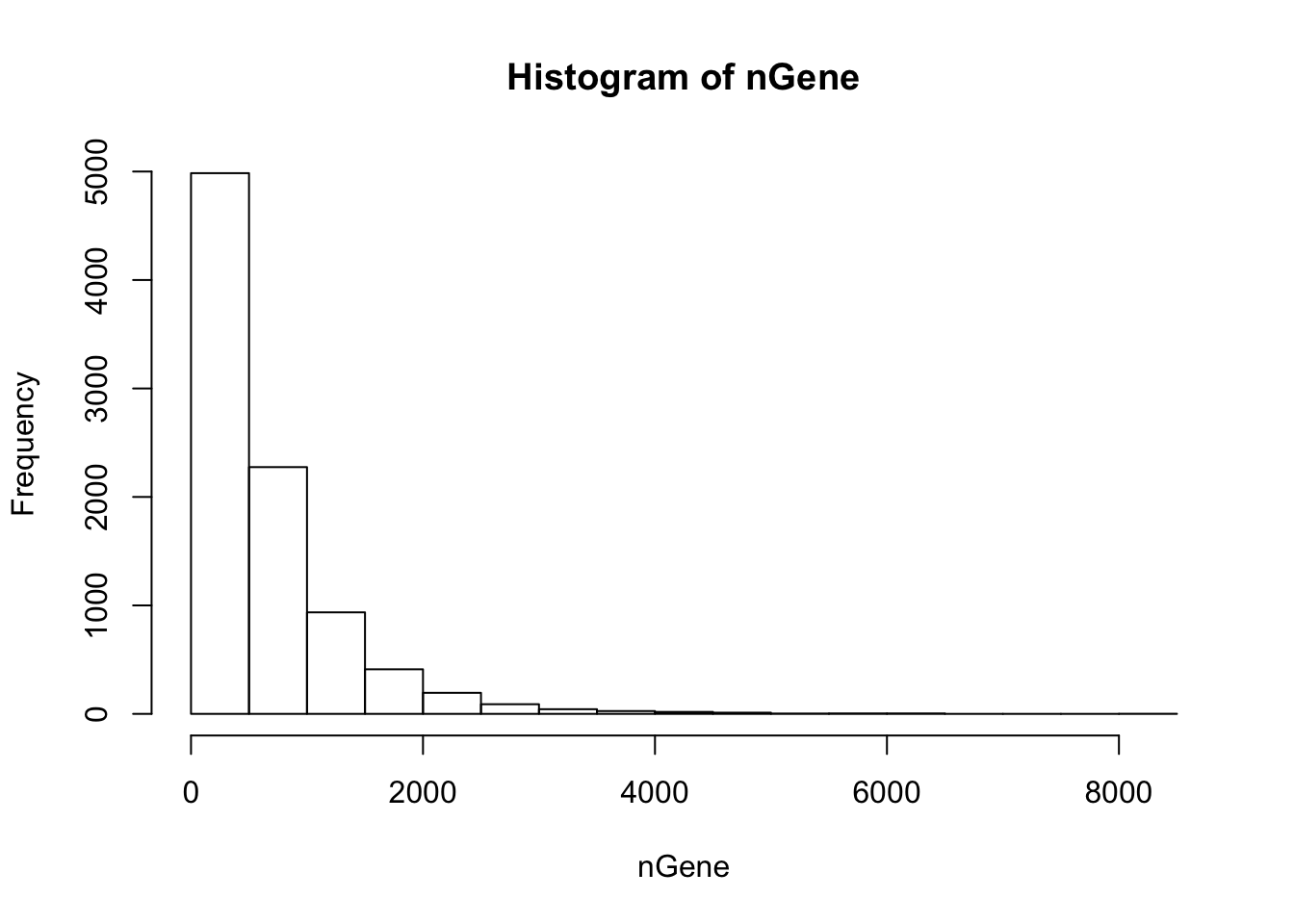
Expand here to see past versions of cell_filter-1.png:
| Version | Author | Date |
|---|---|---|
| f6c8902 | tk382 | 2018-08-29 |
summaryX = cellFilter(X = orig,
genenames = genenames,
minGene = 500,
maxGene = 2000,
maxMitoProp = 0.1)
tmpX = summaryX$X
nUMI = summaryX$nUMI
nGene = summaryX$nGene
percent.mito = summaryX$percent.mito
det.rate = summaryX$det.rate
par(mfrow = c(1,4))
boxplot(nUMI, main='nUMI');
boxplot(nGene, main='nGene');
boxplot(percent.mito, main='mitochondrial gene', ylim=c(0,0.5));
boxplot(det.rate, main='detection rate', ylim=c(0,0.1))
Expand here to see past versions of cell_filter-2.png:
| Version | Author | Date |
|---|---|---|
| f6c8902 | tk382 | 2018-08-29 |
Gene filtering
X = tmpX[rowSums(tmpX) > 0, ]
genenames = genenames[rowSums(tmpX) > 0]
#gene filter by dispersion
disp = dispersion(X, bins = 20)
plot(disp$z ~ disp$genemeans,
xlab = "mean expression",
ylab = "normalized dispersion")
Expand here to see past versions of basic_gene_filtering-1.png:
| Version | Author | Date |
|---|---|---|
| f6c8902 | tk382 | 2018-08-29 |
select = which(abs(disp$z) > 1)
X = X[select, ]
genenames = genenames[select]UMI Normalization
Use quantile-normalization to make the distribution of each cell the same.
nX = quantile_normalize(as.matrix(X))Correct Detection Rate
After normalization, the linear relationship between the first PC and the detection rate usually disappears. The plots show that even without correction, the two are not heavily correlated. So we do not regress out the detection rate.
#take log
logX = as.matrix(log(nX + 1))
#check dependency
out = correct_detection_rate(logX, det.rate)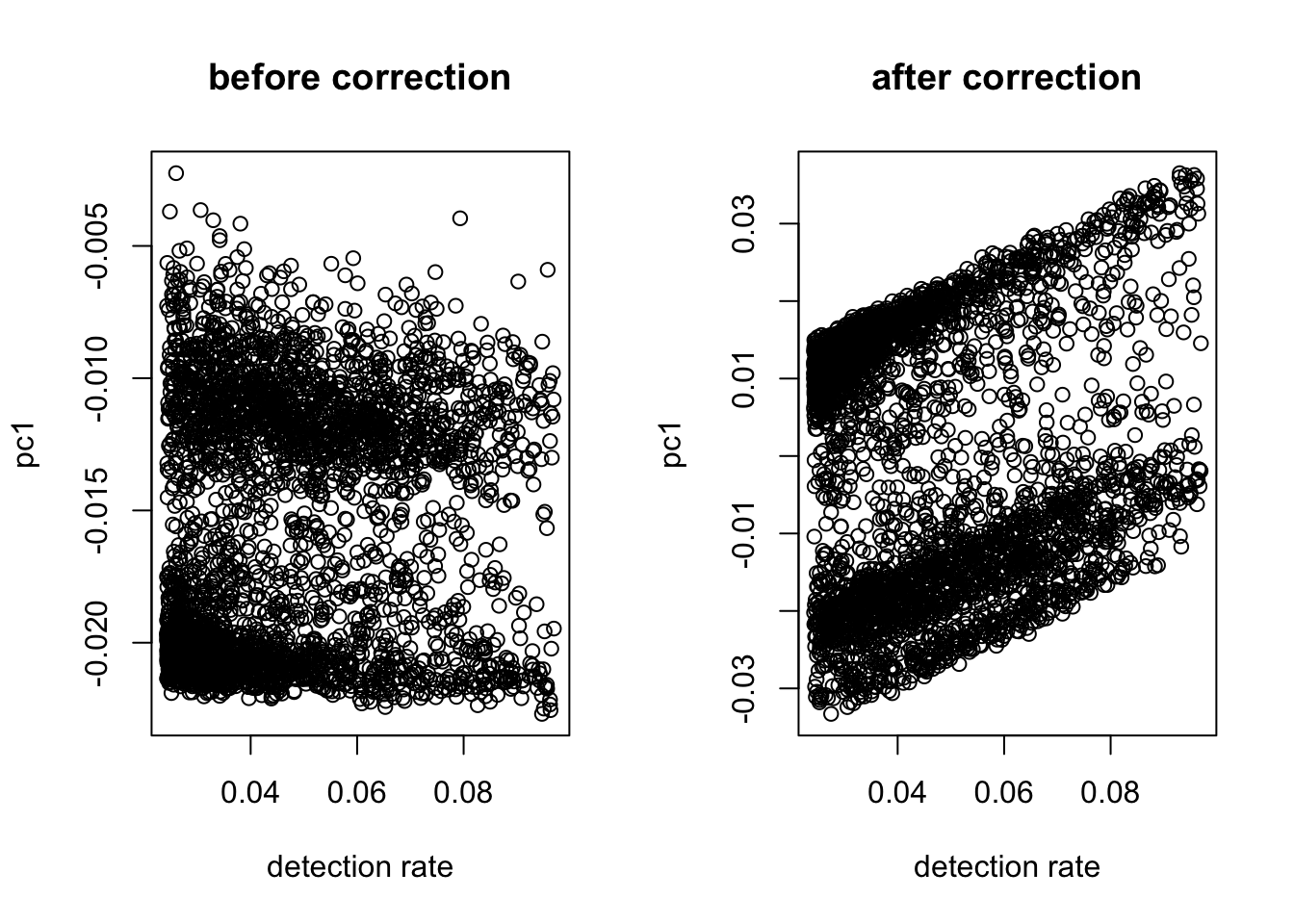
Expand here to see past versions of log_transform_and_det_correction-1.png:
| Version | Author | Date |
|---|---|---|
| f6c8902 | tk382 | 2018-08-29 |
#regress out
# log.cpm = out$residual
log.cpm = logXDimension reduction on the data for visualization.
pc = irlba(log.cpm, 20)
plot(pc$d, ylab = "singular values")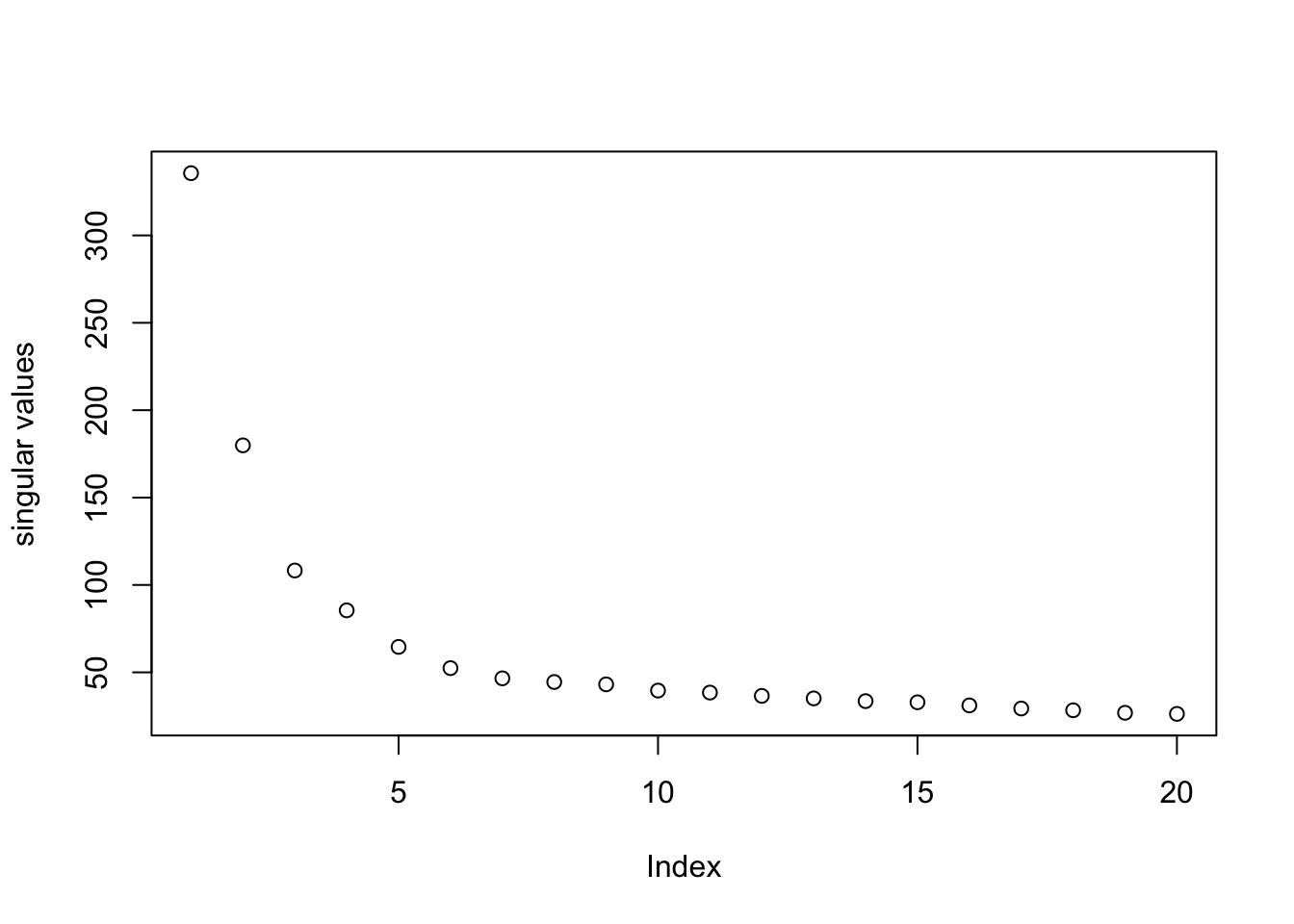
Expand here to see past versions of tsne-1.png:
| Version | Author | Date |
|---|---|---|
| f6c8902 | tk382 | 2018-08-29 |
tsne = Rtsne(pc$v[,1:5], dims=2, perplexity = 100, pca=FALSE)
plot(tsne$Y, xlab = 'tsne1', ylab = 'tsne2', cex = 0.5)
Expand here to see past versions of tsne-2.png:
| Version | Author | Date |
|---|---|---|
| f6c8902 | tk382 | 2018-08-29 |
rm(pc)Run SLSL on the log.cpm matrix.
# out = SLSL(log.cpm, log=FALSE,
# filter = FALSE,
# correct_detection_rate = FALSE,
# klist = c(300,350,400),
# sigmalist = c(1,1.5,2),
# kernel_type = "pearson",
# verbose=FALSE)
# save(out, file = 'dropseq_slsl1.Rdata')
load('analysis/dropseq_slsl1.Rdata')
tab = table(out$result)
plot(tsne$Y, col=rainbow(length(tab))[out$result],
xlab = 'tsne1', ylab='tsne2', main="SLSL", cex = 0.5)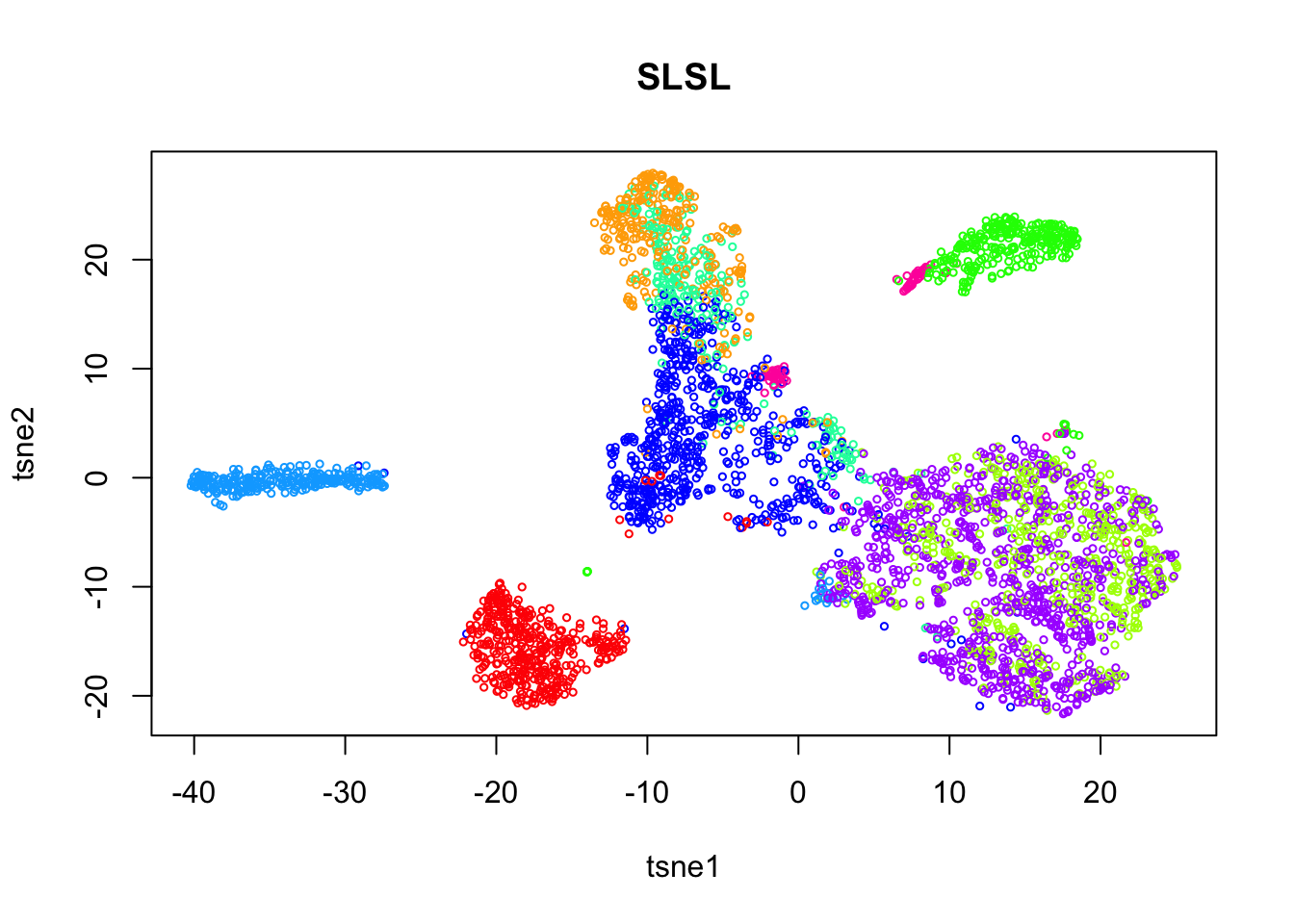
Using SC3 with our gene and cell filter procedure and disabling the biology and filter feature of the function leads to the following result. Using SC3’s default methods lead to the error “distribution of gene expression in cells is too skewed towards 0”.
library(SingleCellExperiment)
library(SC3)
# colnames(X) = paste0("C", 1:ncol(X))
# sce = SingleCellExperiment(
# assays = list(
# counts = X,
# logcounts = as.matrix(log(X+1))
# ),
# colData = colnames(X)
# )
# rowData(sce)$feature_symbol = genenames
# sce = sc3_prepare(sce, kmeans_nstart = 50)
# sce = sc3_estimate_k(sce)
# k = metadata(sce)$sc3$k_estimation
# N = ncol(X)
# sce = sc3(sce, ks=k, biology = FALSE, gene_filter=FALSE,
# kmeans_nstart=10)
# col_data_selected = colData(sce)
# save(col_data_selected, file = "analysis/dropseq_sc3_result.Rdata")
load('analysis/dropseq_sc3_result.Rdata')
plot(tsne$Y, col=rainbow(k)[col_data_selected$sc3_6_clusters],
xlab = 'tsne1', ylab='tsne2', main="SC3", cex = 0.5)ind = which(genenames %in% c('Chat', 'Gad1', 'Gad2', 'Slc17a8', 'Slc6a9', 'Gjd2'))
df = data.frame(tsne1 = tsne$Y[,1], tsne2 = tsne$Y[,2],
Slc17a8 = log.cpm[ind[1], ],
Gad2 = log.cpm[ind[2], ],
Gad1 = log.cpm[ind[3],]
)
g1 = ggplot(df, aes(x=tsne1, y=tsne2, col = Slc17a8)) + geom_point(size=0.1) +
scale_colour_gradient(low="pink", high="black") + guides(color = FALSE) + ggtitle('Slc17a8')
g2 = ggplot(df, aes(x=tsne1, y=tsne2, col = Gad2)) + geom_point(size=0.1) +
scale_colour_gradient(low="pink", high="black") + guides(color = FALSE) + ggtitle('Gad2')
g3 = ggplot(df, aes(x=tsne1, y=tsne2, col = Gad1)) + geom_point(size=0.1) +
scale_colour_gradient(low="pink", high="black") + guides(color = FALSE) + ggtitle('Gad1')
grid.arrange(g1,g2,g3, nrow=1)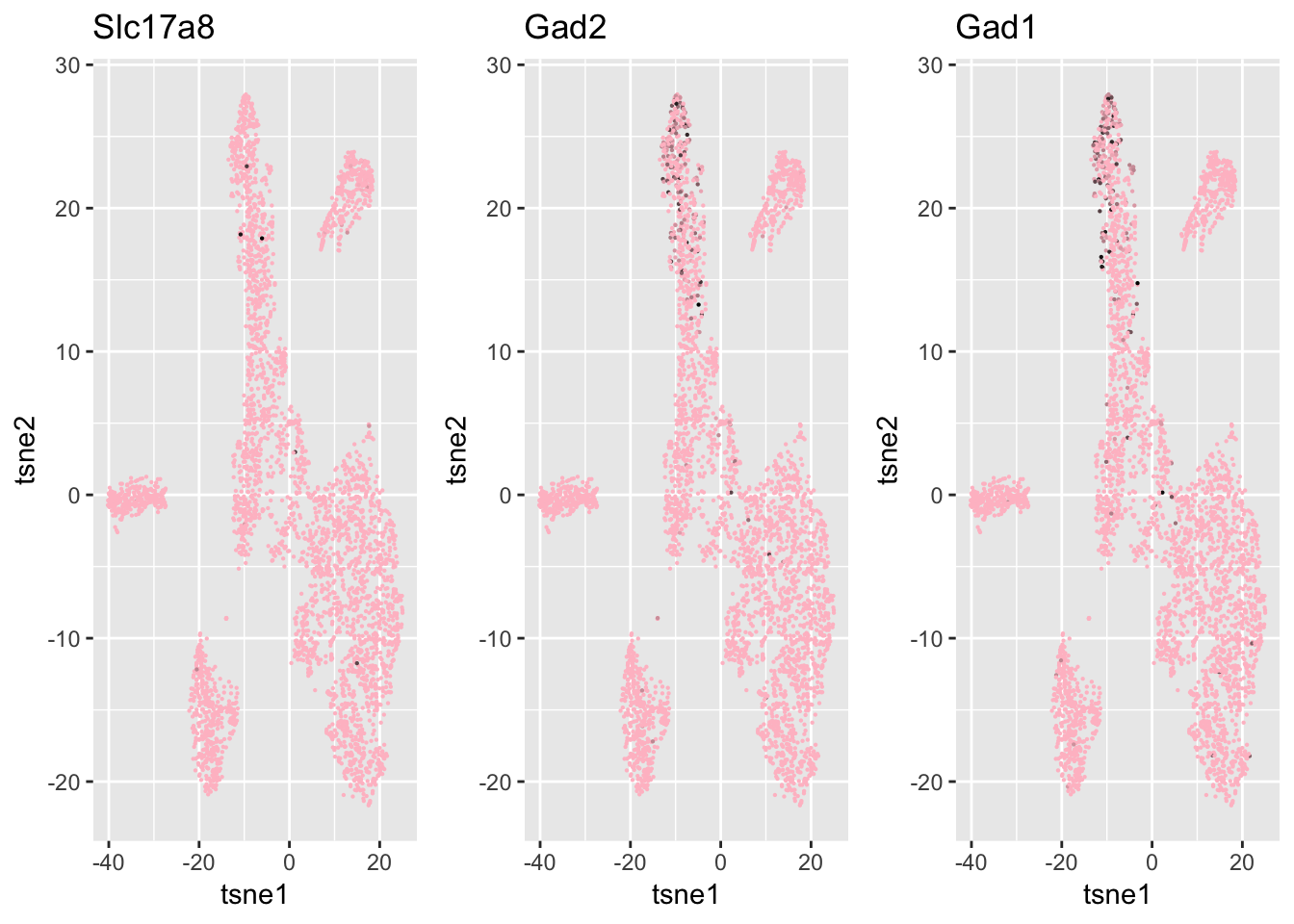
Expand here to see past versions of markers-1.png:
| Version | Author | Date |
|---|---|---|
| f6c8902 | tk382 | 2018-08-29 |
Session information
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Sierra 10.12.5
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] parallel stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] bindrcpp_0.2.2 data.table_1.11.4
[3] gridExtra_2.3 gdata_2.18.0
[5] stargazer_5.2.2 abind_1.4-5
[7] broom_0.5.0 gplots_3.0.1
[9] diceR_0.5.1 Rtsne_0.13
[11] igraph_1.2.2 scatterplot3d_0.3-41
[13] pracma_2.1.4 fossil_0.3.7
[15] shapefiles_0.7 foreign_0.8-71
[17] maps_3.3.0 sp_1.3-1
[19] caret_6.0-80 lattice_0.20-35
[21] reshape_0.8.7 dplyr_0.7.6
[23] ggplot2_3.0.0 irlba_2.3.2
[25] Matrix_1.2-14 quadprog_1.5-5
[27] inline_0.3.15 matrixStats_0.54.0
[29] SCNoisyClustering_0.1.0
loaded via a namespace (and not attached):
[1] nlme_3.1-137 bitops_1.0-6
[3] lubridate_1.7.4 dimRed_0.1.0
[5] rprojroot_1.3-2 tools_3.5.1
[7] backports_1.1.2 R6_2.2.2
[9] KernSmooth_2.23-15 rpart_4.1-13
[11] lazyeval_0.2.1 colorspace_1.3-2
[13] nnet_7.3-12 withr_2.1.2
[15] tidyselect_0.2.4 compiler_3.5.1
[17] git2r_0.23.0 labeling_0.3
[19] caTools_1.17.1.1 scales_0.5.0
[21] sfsmisc_1.1-2 DEoptimR_1.0-8
[23] robustbase_0.93-2 stringr_1.3.1
[25] digest_0.6.15 rmarkdown_1.10
[27] R.utils_2.6.0 pkgconfig_2.0.1
[29] htmltools_0.3.6 rlang_0.2.1
[31] ddalpha_1.3.4 bindr_0.1.1
[33] gtools_3.8.1 mclust_5.4.1
[35] ModelMetrics_1.1.0 R.oo_1.22.0
[37] magrittr_1.5 Rcpp_0.12.18
[39] munsell_0.5.0 R.methodsS3_1.7.1
[41] stringi_1.2.4 whisker_0.3-2
[43] yaml_2.2.0 MASS_7.3-50
[45] plyr_1.8.4 recipes_0.1.3
[47] grid_3.5.1 pls_2.6-0
[49] crayon_1.3.4 splines_3.5.1
[51] knitr_1.20 pillar_1.3.0
[53] reshape2_1.4.3 codetools_0.2-15
[55] stats4_3.5.1 CVST_0.2-2
[57] magic_1.5-8 glue_1.3.0
[59] evaluate_0.11 RcppArmadillo_0.8.600.0.0
[61] foreach_1.4.4 gtable_0.2.0
[63] purrr_0.2.5 tidyr_0.8.1
[65] kernlab_0.9-26 assertthat_0.2.0
[67] DRR_0.0.3 gower_0.1.2
[69] prodlim_2018.04.18 class_7.3-14
[71] survival_2.42-6 geometry_0.3-6
[73] timeDate_3043.102 RcppRoll_0.3.0
[75] tibble_1.4.2 iterators_1.0.10
[77] workflowr_1.1.1 lava_1.6.2
[79] ipred_0.9-6 This reproducible R Markdown analysis was created with workflowr 1.1.1