Shrink R squared - examples
Dongyue Xie
2019-01-21
Last updated: 2019-02-10
workflowr checks: (Click a bullet for more information)-
✔ R Markdown file: up-to-date
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
-
✔ Environment: empty
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
-
✔ Seed:
set.seed(20180501)The command
set.seed(20180501)was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible. -
✔ Session information: recorded
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
-
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.✔ Repository version: d255e06
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can usewflow_publishorwflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.Ignored files: Ignored: .DS_Store Ignored: .Rhistory Ignored: .Rproj.user/ Ignored: data/.DS_Store Untracked files: Untracked: analysis/chipexoeg.Rmd Untracked: analysis/efsd.Rmd Untracked: analysis/talk1011.Rmd Untracked: data/chipexo_examples/ Untracked: data/chipseq_examples/ Untracked: talk.Rmd Untracked: talk.pdf Unstaged changes: Modified: analysis/binomial.Rmd Modified: analysis/fda.Rmd Modified: analysis/index.Rmd Modified: analysis/r2.Rmd Modified: analysis/sigma.Rmd
Expand here to see past versions:
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | d255e06 | Dongyue Xie | 2019-02-10 | wflow_publish(“analysis/r2b.Rmd”) |
| html | c6f9a91 | Dongyue Xie | 2019-02-10 | Build site. |
| Rmd | 7e08c59 | Dongyue Xie | 2019-02-10 | wflow_publish(“analysis/r2b.Rmd”) |
| html | c7f4704 | Dongyue Xie | 2019-01-27 | Build site. |
| Rmd | a1a64c5 | Dongyue Xie | 2019-01-27 | wflow_publish(“analysis/r2b.Rmd”) |
| html | 9ce257c | Dongyue Xie | 2019-01-27 | Build site. |
| Rmd | d6bcb1a | Dongyue Xie | 2019-01-27 | wflow_publish(“analysis/r2b.Rmd”) |
| html | 97b73fc | Dongyue Xie | 2019-01-22 | Build site. |
| Rmd | b5029e2 | Dongyue Xie | 2019-01-22 | wflow_publish(“analysis/r2b.Rmd”) |
| html | ad12c40 | Dongyue Xie | 2019-01-22 | Build site. |
| Rmd | 11f83fb | Dongyue Xie | 2019-01-22 | wflow_publish(“analysis/r2b.Rmd”) |
For the method used in these examples, see here
True \(R^2\) is defined as \(R^2=\frac{var(X\beta)}{var(y)}=\frac{var(y)-\sigma^2}{var(y)}=1-\frac{\sigma^2}{var(y)}=1-\frac{\sigma^2}{\sigma^2+var(X\beta)}\)
Ajusted R^2: \(1-\frac{\sum(y_i-\hat y_i)^2/(n-p-1)}{\sum(y_i-\bar y)^2/(n-1)}\)
Shrunk adjusted R^2: use
fashshrinking \(fash.output=\log(\frac{\sum(y_i-\hat y_i)^2/(n-p-1)}{\sum(y_i-\bar y)^2/(n-1)})\) then shrunk adjusted R^2 is \(1-\exp(fash.output)\)Shrunk R^2: use
fashshrinking \(fash.output=\log(\frac{\sum(y_i-\hat y_i)^2/(n-1)}{\sum(y_i-\bar y)^2/(n-1)})\) then shrunk adjusted R^2 is \(1-\exp(fash.output)\)Another Shrunk R^2: shrink all \(\betas\) using
ash, obtain posterior means then calculate \(\hat\sigma^2\) then obtain \(\frac{var(X\hat\beta)}{var(\hat\beta)+\hat\sigma^2}\).
(note: this is the old idea which introduces bias to R^2 when multiplying \(\frac{n-p-1}{n-1}\) so I discard this method.)(Shrunk R^2 = \(1 - \exp(fash.output)*\frac{n-p-1}{n-1}\), because \(R^2=1-\frac{\sum(y_i-\hat y_i)^2}{\sum(y_i-\bar y)^2}=1-\frac{\sum(y_i-\hat y_i)^2/(n-p-1)}{\sum(y_i-\bar y)^2/(n-1)}*\frac{n-p-1}{n-1}=1-(1-adjR^2)*\frac{n-p-1}{n-1}\))
R Function
R function for shrinking adjusted R/ R squared:
library(ashr)
#'@param R2: R squared from linear regression model fit
#'@param n: sample size
#'@param p: the number of covariates
#'@output shrunk R squared.
ash_ar2=function(R2,n,p){
df1=n-p-1
df2=n-1
log.ratio=log((1-R2)/(df1)*(df2))
shrink.log.ratio=ash(log.ratio,1,lik=lik_logF(df1=df1,df2=df2))$result$PosteriorMean
ar2=1-exp(shrink.log.ratio)
return(ar2)
}
ash_r2=function(R2,n,p){
df1=n-1
df2=n-1
log.ratio=log(1-R2)
shrink.log.ratio=ash(log.ratio,1,lik=lik_logF(df1=df1,df2=df2))$result$PosteriorMean
r2=1-exp(shrink.log.ratio)
return(r2)
}Compare Shrunk \(R^2\) with True \(R2\).
Assume linear model \(y=X\beta+\epsilon\) where \(\epsilon\sim N(0,\sigma^2I)\)
- n=100, p=5. Each cordinate of \(\beta\) ranges from 0 to 1, for example \(\beta=(0,0,0,0,0)\),…,\(\beta=(0.1,0.1,0.1,0.1,0.1)\),…, \(\beta=(1,1,1,1,1)\) etc.
If I generate X from Uniform(0,1), then fash shirnks all \(R^2\) to 0. If generate X from Uniform(0,2), then it does not
Uniform(0,1):
set.seed(1234)
n=100
p=5
R2=c()
R2a=c()
trueR2=c()
beta.list=seq(0,1,length.out = 100)
X=matrix(runif(n*(p),0,1),n,p)
for (i in 1:length(beta.list)) {
beta=rep(beta.list[i],p)
y=X%*%beta+rnorm(n)
datax=data.frame(X=X,y=y)
mod=lm(y~.,datax)
mod.sy=summary(mod)
R2[i]=mod.sy$r.squared
R2a[i]=mod.sy$adj.r.squared
trueR2[i]=var(X%*%beta)/(1+var(X%*%beta))
}
R2s=ash_r2(R2,n,p)
R2as=ash_ar2(R2,n,p)
plot(beta.list,R2,ylim=range(c(R2,R2a,R2s,R2as,trueR2)),main='',xlab='beta',ylab='',type='l')
lines(beta.list,R2a,col=2)
lines(beta.list,R2as,col=3)
lines(beta.list,R2s,col=4)
lines(beta.list,trueR2,col='grey80')
abline(h=0,lty=2)
legend('topleft',c('R^2','Adjusted R^2','Adjusted R^2 fash','R^2 fash','True R^2'),lty=c(1,1,1,1,1),col=c(1,2,3,4,'grey80'))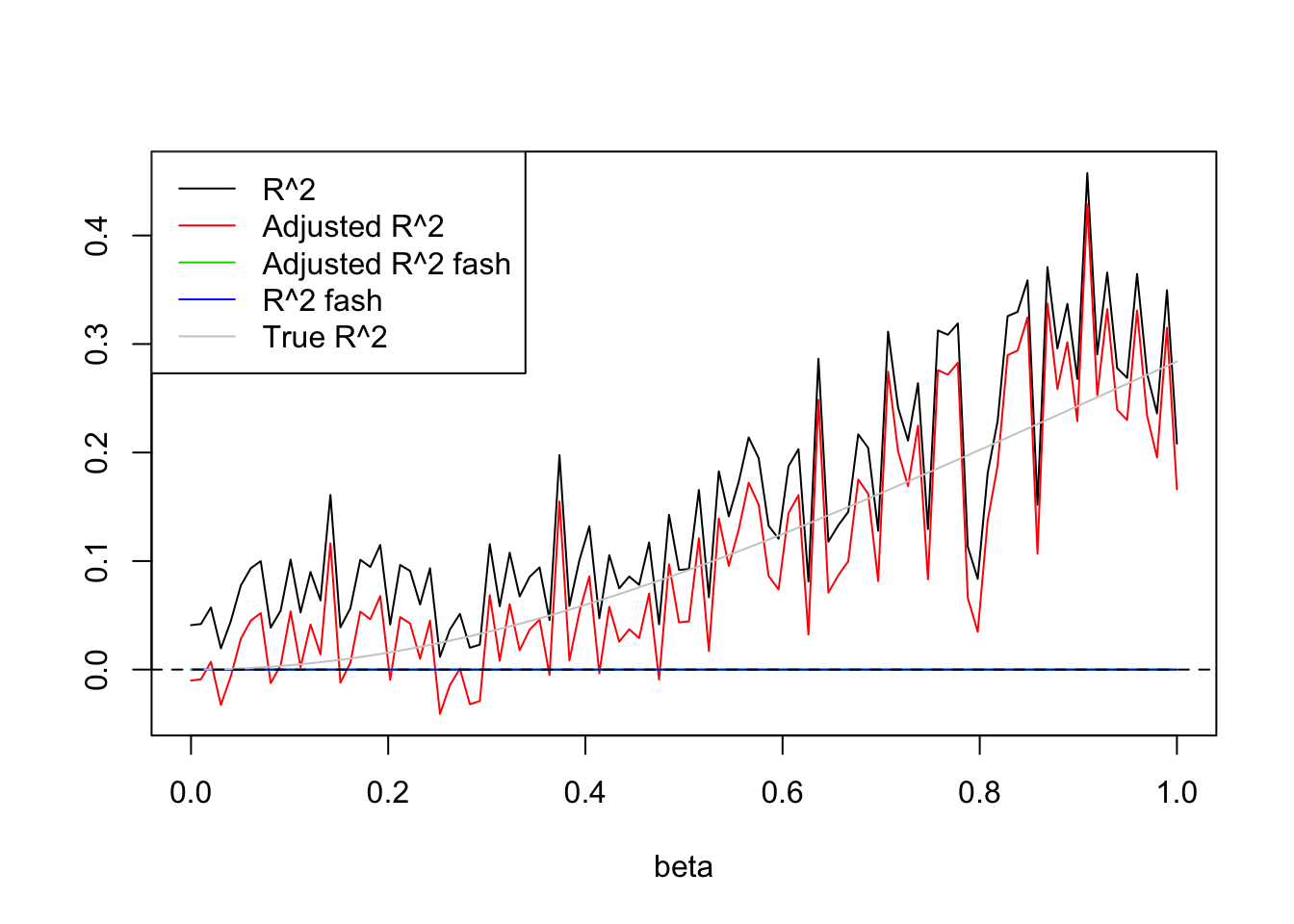
Expand here to see past versions of unnamed-chunk-2-1.png:
| Version | Author | Date |
|---|---|---|
| c6f9a91 | Dongyue Xie | 2019-02-10 |
| 9ce257c | Dongyue Xie | 2019-01-27 |
plot(trueR2,R2,type='l',ylim=range(c(R2,R2a,R2s,R2as,trueR2)))
lines(trueR2,R2a,col=2)
lines(trueR2,R2as,col=3)
lines(trueR2,R2s,col=4)
lines(trueR2,trueR2,col='grey80')
legend('topleft',c('R^2','Adjusted R^2','Adjusted R^2 fash','R^2 fash'),lty=c(1,1,1,1),col=c(1,2,3,4))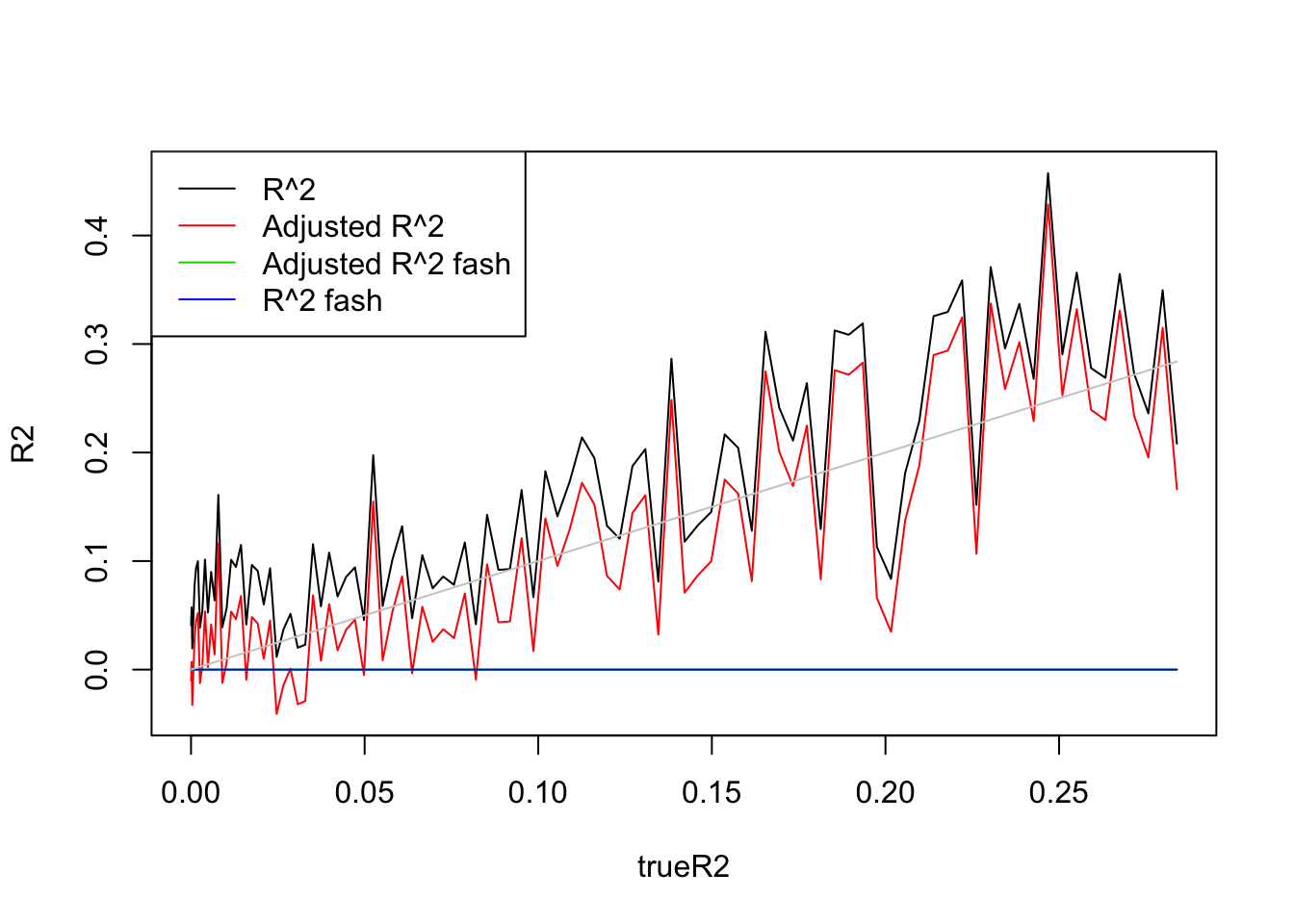
Expand here to see past versions of unnamed-chunk-2-2.png:
| Version | Author | Date |
|---|---|---|
| c6f9a91 | Dongyue Xie | 2019-02-10 |
| 9ce257c | Dongyue Xie | 2019-01-27 |
Uniform(0,3):
set.seed(1234)
n=100
p=5
R2=c()
R2a=c()
trueR2=c()
beta.list=seq(0,1,length.out = 100)
X=matrix(runif(n*(p),0,3),n,p)
for (i in 1:length(beta.list)) {
beta=rep(beta.list[i],p)
y=X%*%beta+rnorm(n)
datax=data.frame(X=X,y=y)
mod=lm(y~.,datax)
mod.sy=summary(mod)
R2[i]=mod.sy$r.squared
R2a[i]=mod.sy$adj.r.squared
trueR2[i]=var(X%*%beta)/(1+var(X%*%beta))
}
R2s=ash_r2(R2,n,p)
R2as=ash_ar2(R2,n,p)
plot(beta.list,R2,ylim=range(c(R2,R2a,R2s,R2as,trueR2)),main='',xlab='beta',ylab='',type='l')
lines(beta.list,R2a,col=2)
lines(beta.list,R2as,col=3)
lines(beta.list,R2s,col=4)
lines(beta.list,trueR2,col='grey80')
abline(h=0,lty=2)
legend('topleft',c('R^2','Adjusted R^2','Adjusted R^2 fash','R^2 fash','True R^2'),lty=c(1,1,1,1,1),col=c(1,2,3,4,'grey80'))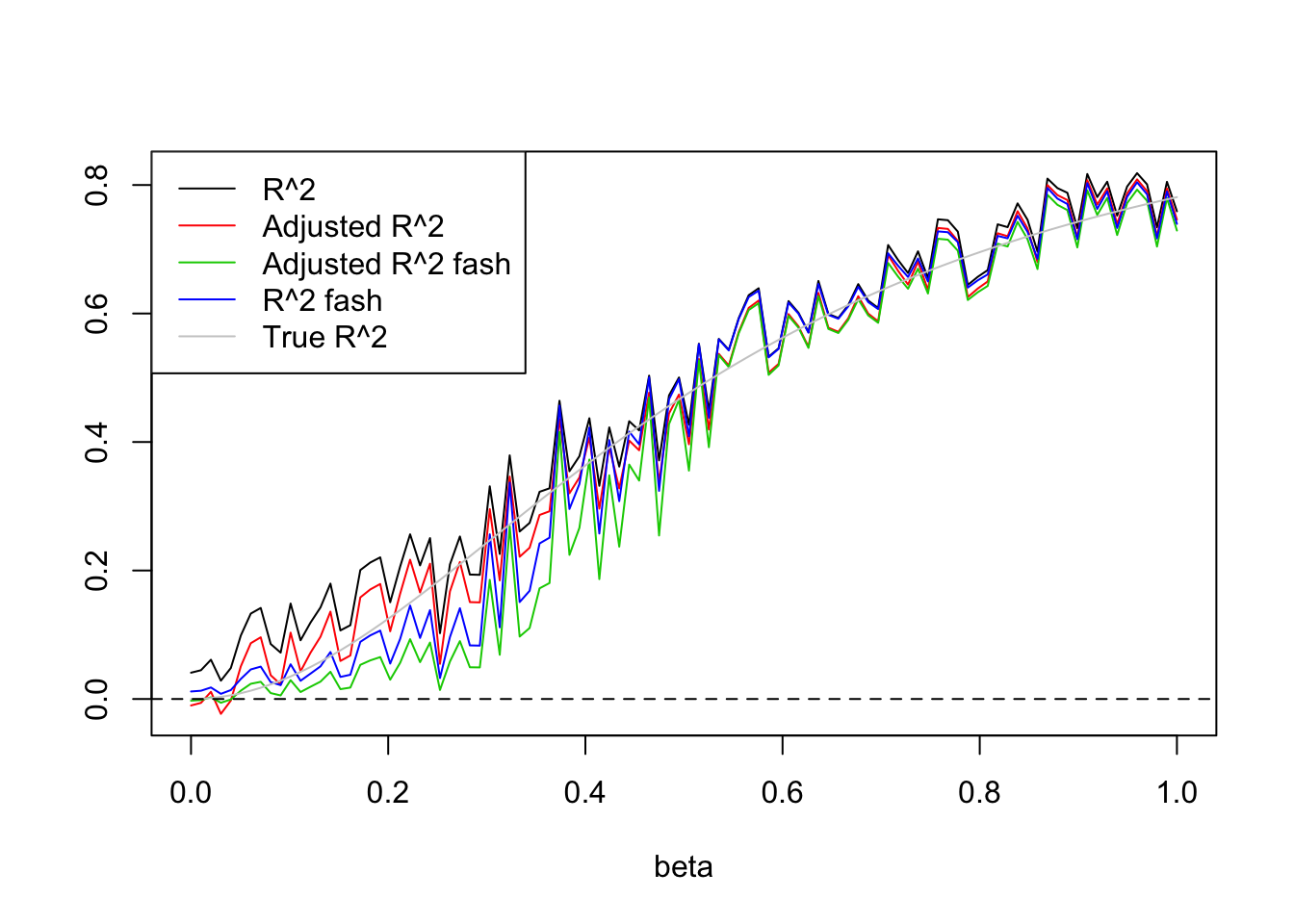
Expand here to see past versions of unnamed-chunk-3-1.png:
| Version | Author | Date |
|---|---|---|
| c6f9a91 | Dongyue Xie | 2019-02-10 |
| 9ce257c | Dongyue Xie | 2019-01-27 |
plot(trueR2,R2,type='l',ylim=range(c(R2,R2a,R2s,R2as,trueR2)))
lines(trueR2,R2a,col=2)
lines(trueR2,R2as,col=3)
lines(trueR2,R2s,col=4)
lines(trueR2,trueR2,col='grey80')
legend('topleft',c('R^2','Adjusted R^2','Adjusted R^2 fash','R^2 fash'),lty=c(1,1,1,1),col=c(1,2,3,4))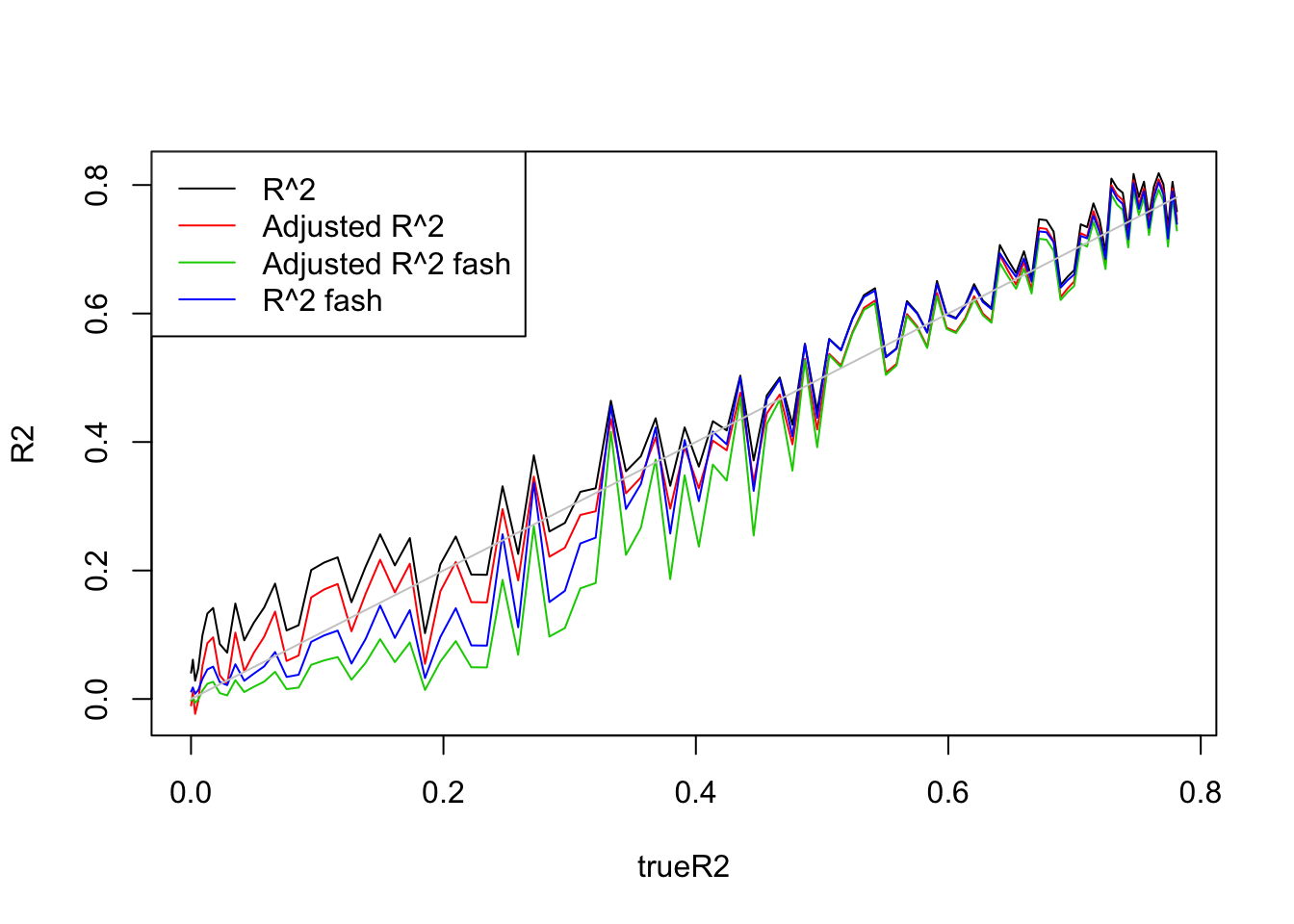
Expand here to see past versions of unnamed-chunk-3-2.png:
| Version | Author | Date |
|---|---|---|
| c6f9a91 | Dongyue Xie | 2019-02-10 |
| 9ce257c | Dongyue Xie | 2019-01-27 |
Uniform(0,5):
set.seed(1234)
n=100
p=5
R2=c()
R2a=c()
trueR2=c()
beta.list=seq(0,1,length.out = 100)
X=matrix(runif(n*(p),0,5),n,p)
for (i in 1:length(beta.list)) {
beta=rep(beta.list[i],p)
y=X%*%beta+rnorm(n)
datax=data.frame(X=X,y=y)
mod=lm(y~.,datax)
mod.sy=summary(mod)
R2[i]=mod.sy$r.squared
R2a[i]=mod.sy$adj.r.squared
trueR2[i]=var(X%*%beta)/(1+var(X%*%beta))
}
R2s=ash_r2(R2,n,p)
R2as=ash_ar2(R2,n,p)
plot(beta.list,R2,ylim=range(c(R2,R2a,R2s,R2as,trueR2)),main='',xlab='beta',ylab='',type='l')
lines(beta.list,R2a,col=2)
lines(beta.list,R2as,col=3)
lines(beta.list,R2s,col=4)
lines(beta.list,trueR2,col='grey80')
abline(h=0,lty=2)
legend('topleft',c('R^2','Adjusted R^2','Adjusted R^2 fash','R^2 fash','True R^2'),lty=c(1,1,1,1,1),col=c(1,2,3,4,'grey80'))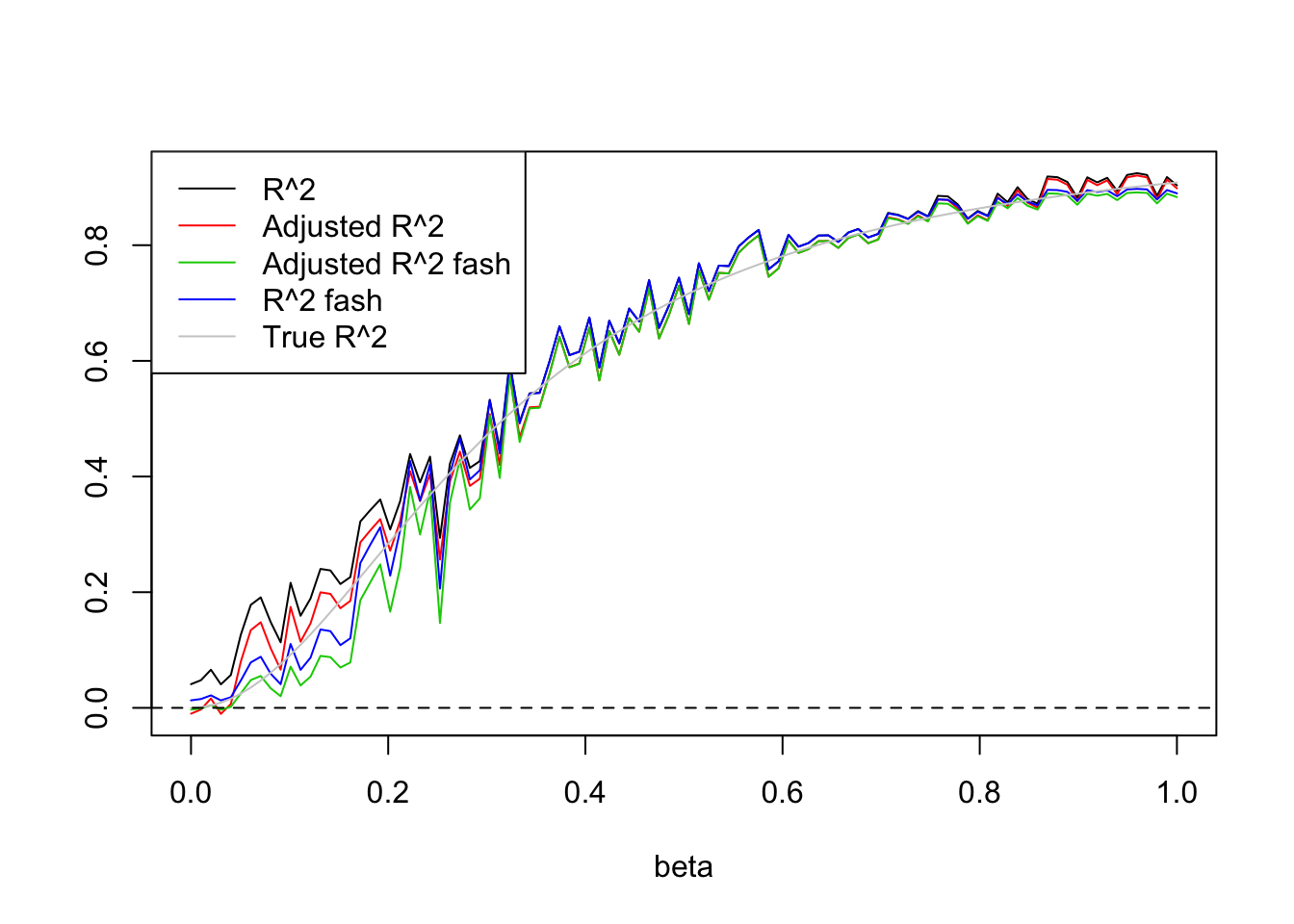
Expand here to see past versions of unnamed-chunk-4-1.png:
| Version | Author | Date |
|---|---|---|
| c6f9a91 | Dongyue Xie | 2019-02-10 |
| 9ce257c | Dongyue Xie | 2019-01-27 |
plot(trueR2,R2,type='l',ylim=range(c(R2,R2a,R2s,R2as,trueR2)))
lines(trueR2,R2a,col=2)
lines(trueR2,R2as,col=3)
lines(trueR2,R2s,col=4)
lines(trueR2,trueR2,col='grey80')
legend('topleft',c('R^2','Adjusted R^2','Adjusted R^2 fash','R^2 fash'),lty=c(1,1,1,1),col=c(1,2,3,4))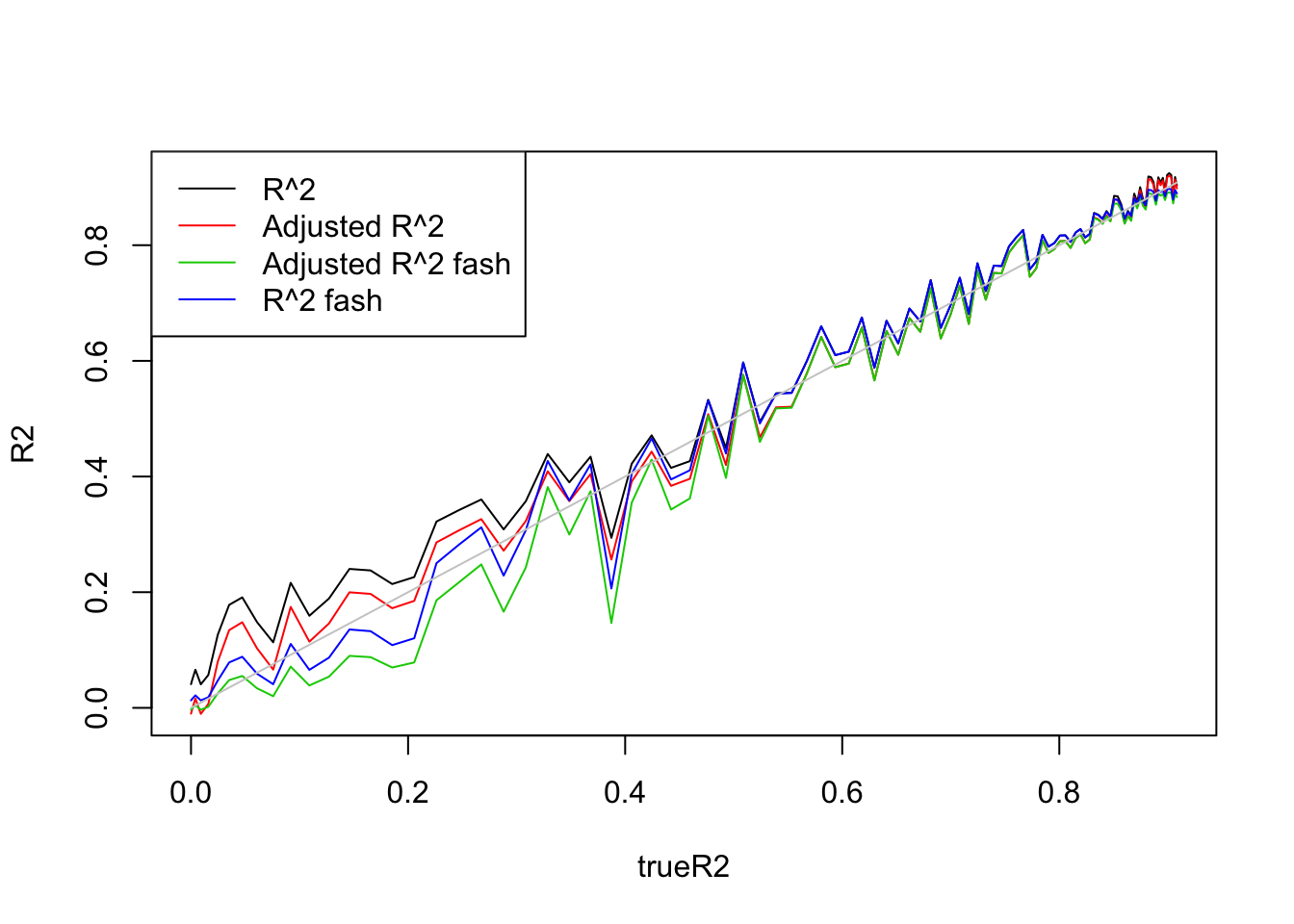
Expand here to see past versions of unnamed-chunk-4-2.png:
| Version | Author | Date |
|---|---|---|
| c6f9a91 | Dongyue Xie | 2019-02-10 |
| 9ce257c | Dongyue Xie | 2019-01-27 |
- Increase \(p\) to 20.
This time, if I generate X from Uniform(0,1), then fash does not shirnk all \(R^2\) to 0.
Uniform(0,1):
set.seed(1234)
n=100
p=20
R2=c()
R2a=c()
trueR2=c()
beta.list=seq(0,1,length.out = 100)
X=matrix(runif(n*(p),0,1),n,p)
for (i in 1:length(beta.list)) {
beta=rep(beta.list[i],p)
y=X%*%beta+rnorm(n)
datax=data.frame(X=X,y=y)
mod=lm(y~.,datax)
mod.sy=summary(mod)
R2[i]=mod.sy$r.squared
R2a[i]=mod.sy$adj.r.squared
trueR2[i]=var(X%*%beta)/(1+var(X%*%beta))
}
R2s=ash_r2(R2,n,p)
R2as=ash_ar2(R2,n,p)
plot(beta.list,R2,ylim=range(c(R2,R2a,R2s,R2as,trueR2)),main='',xlab='beta',ylab='',type='l')
lines(beta.list,R2a,col=2)
lines(beta.list,R2as,col=3)
lines(beta.list,R2s,col=4)
lines(beta.list,trueR2,col='grey80')
abline(h=0,lty=2)
legend('topleft',c('R^2','Adjusted R^2','Adjusted R^2 fash','R^2 fash','True R^2'),lty=c(1,1,1,1,1),col=c(1,2,3,4,'grey80'))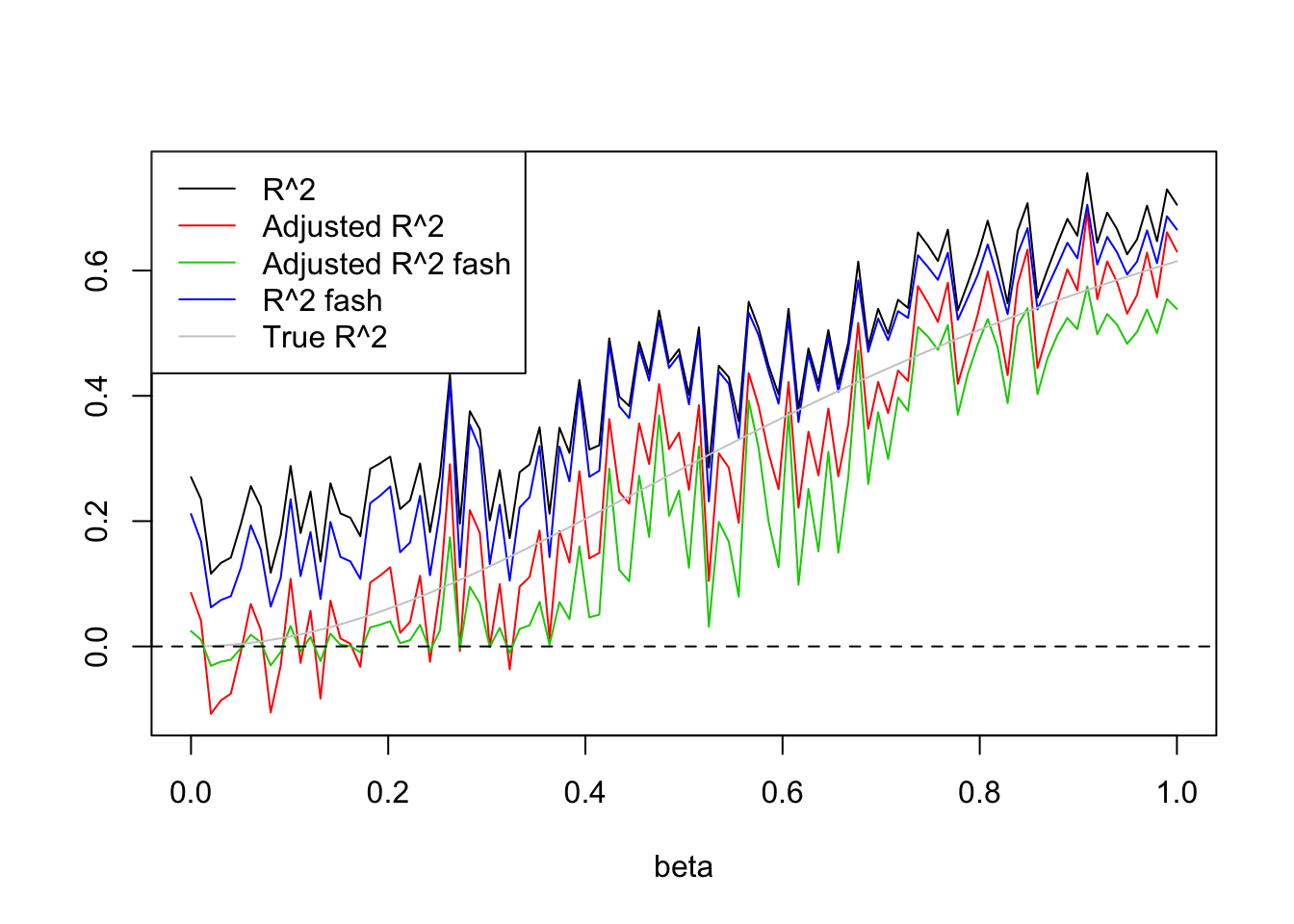
Expand here to see past versions of unnamed-chunk-5-1.png:
| Version | Author | Date |
|---|---|---|
| c6f9a91 | Dongyue Xie | 2019-02-10 |
| c7f4704 | Dongyue Xie | 2019-01-27 |
| 9ce257c | Dongyue Xie | 2019-01-27 |
plot(trueR2,R2,type='l',ylim=range(c(R2,R2a,R2s,R2as,trueR2)))
lines(trueR2,R2a,col=2)
lines(trueR2,R2as,col=3)
lines(trueR2,R2s,col=4)
lines(trueR2,trueR2,col='grey80')
legend('topleft',c('R^2','Adjusted R^2','Adjusted R^2 fash','R^2 fash'),lty=c(1,1,1,1),col=c(1,2,3,4))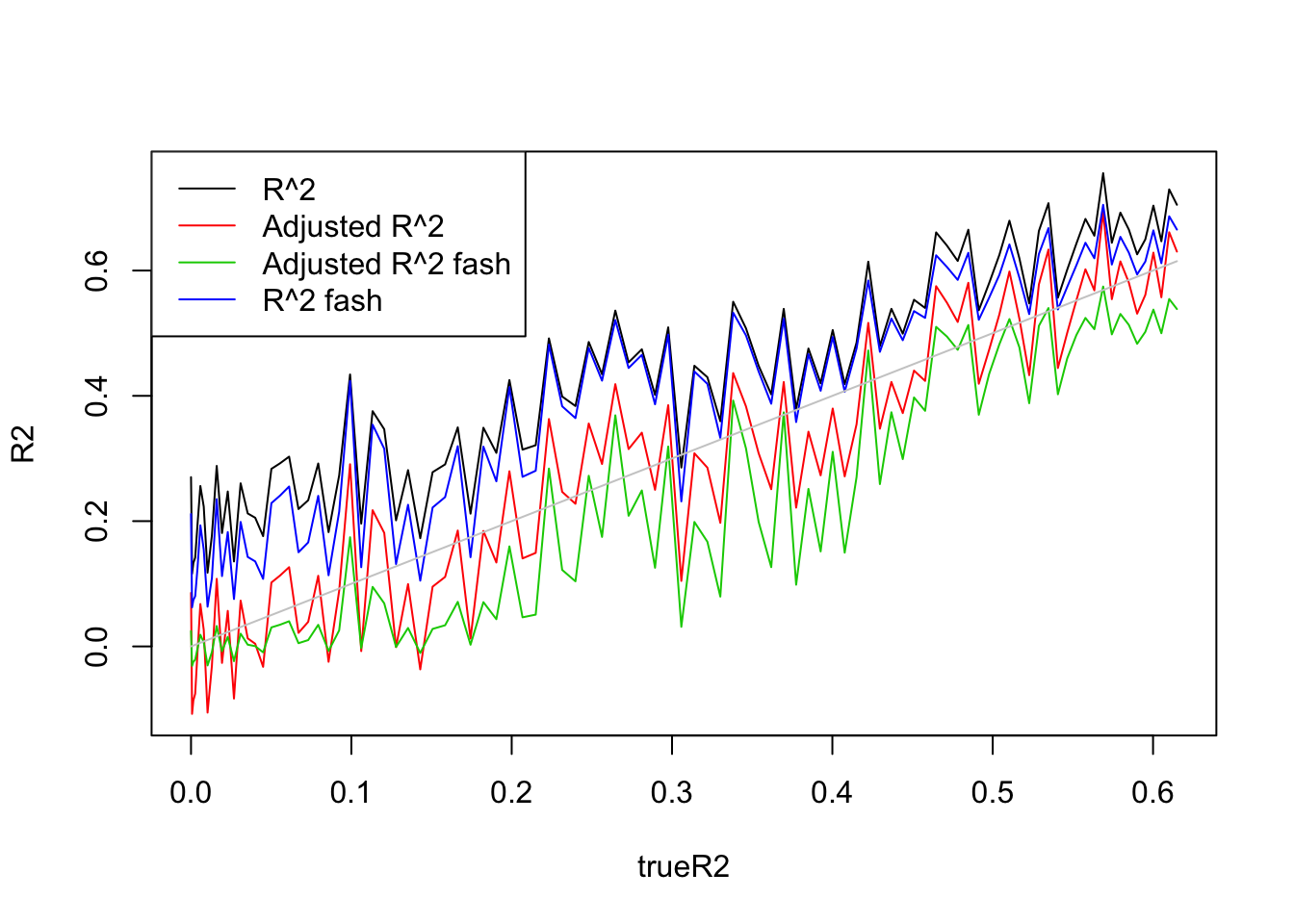
Expand here to see past versions of unnamed-chunk-5-2.png:
| Version | Author | Date |
|---|---|---|
| c6f9a91 | Dongyue Xie | 2019-02-10 |
| c7f4704 | Dongyue Xie | 2019-01-27 |
| 9ce257c | Dongyue Xie | 2019-01-27 |
- \(n=30, p=3\).
This time, I have to generate X from at least Uniform(0,3) to avoid over-shrinkage of fash. Here, I tried Uniform(0,5)
Uniform(0,5):
set.seed(1234)
n=30
p=3
R2=c()
R2a=c()
trueR2=c()
beta.list=seq(0,1,length.out = 100)
X=matrix(runif(n*(p),0,5),n,p)
for (i in 1:length(beta.list)) {
beta=rep(beta.list[i],p)
y=X%*%beta+rnorm(n)
datax=data.frame(X=X,y=y)
mod=lm(y~.,datax)
mod.sy=summary(mod)
R2[i]=mod.sy$r.squared
R2a[i]=mod.sy$adj.r.squared
trueR2[i]=var(X%*%beta)/(1+var(X%*%beta))
}
R2s=ash_r2(R2,n,p)
R2as=ash_ar2(R2,n,p)
plot(beta.list,R2,ylim=range(c(R2,R2a,R2s,R2as,trueR2)),main='',xlab='beta',ylab='',type='l')
lines(beta.list,R2a,col=2)
lines(beta.list,R2as,col=3)
lines(beta.list,R2s,col=4)
lines(beta.list,trueR2,col='grey80')
abline(h=0,lty=2)
legend('topleft',c('R^2','Adjusted R^2','Adjusted R^2 fash','R^2 fash','True R^2'),lty=c(1,1,1,1,1),col=c(1,2,3,4,'grey80'))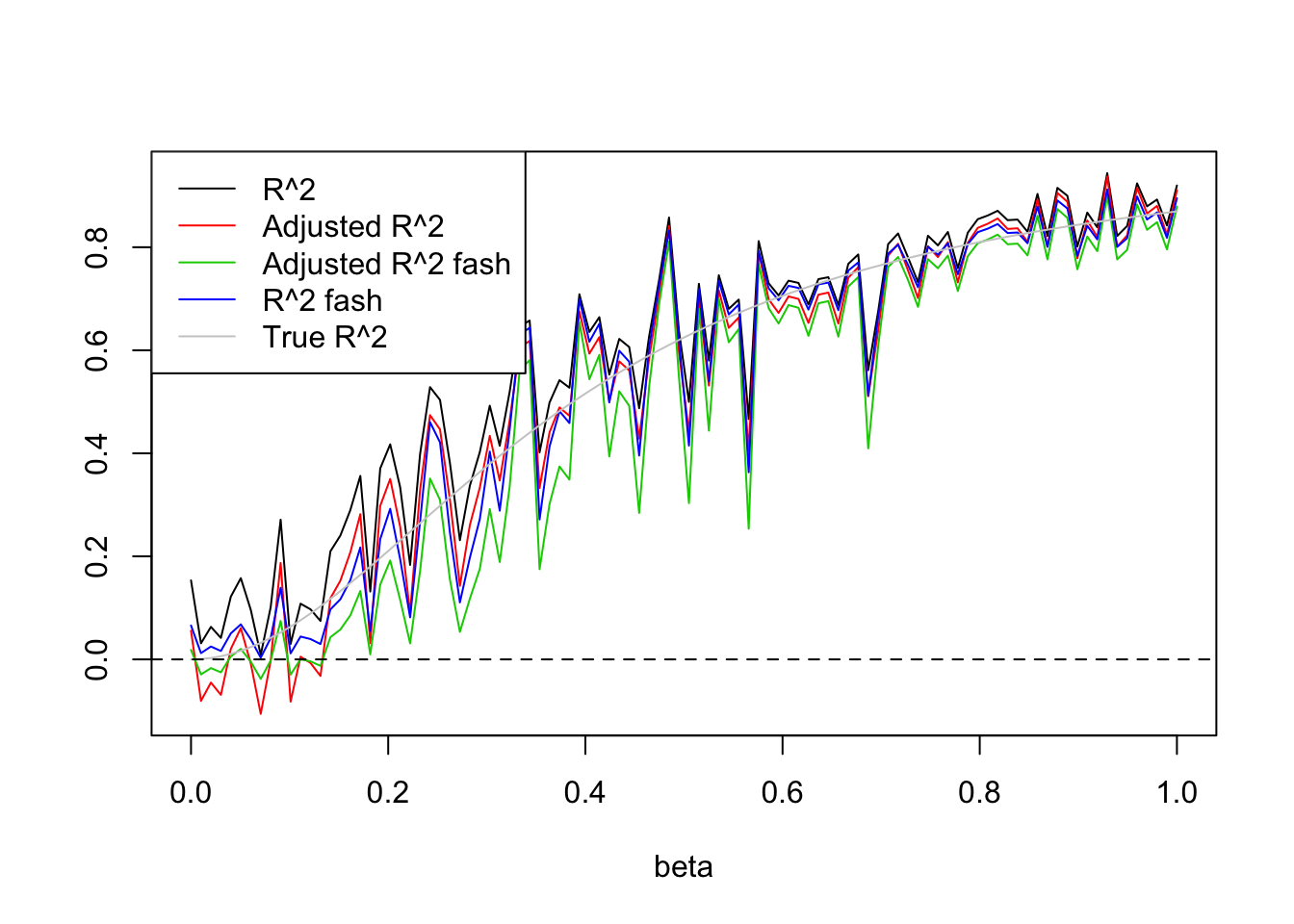
Expand here to see past versions of unnamed-chunk-6-1.png:
| Version | Author | Date |
|---|---|---|
| c6f9a91 | Dongyue Xie | 2019-02-10 |
| c7f4704 | Dongyue Xie | 2019-01-27 |
| 9ce257c | Dongyue Xie | 2019-01-27 |
plot(trueR2,R2,type='l',ylim=range(c(R2,R2a,R2s,R2as,trueR2)))
lines(trueR2,R2a,col=2)
lines(trueR2,R2as,col=3)
lines(trueR2,R2s,col=4)
lines(trueR2,trueR2,col='grey80')
legend('topleft',c('R^2','Adjusted R^2','Adjusted R^2 fash','R^2 fash'),lty=c(1,1,1,1),col=c(1,2,3,4))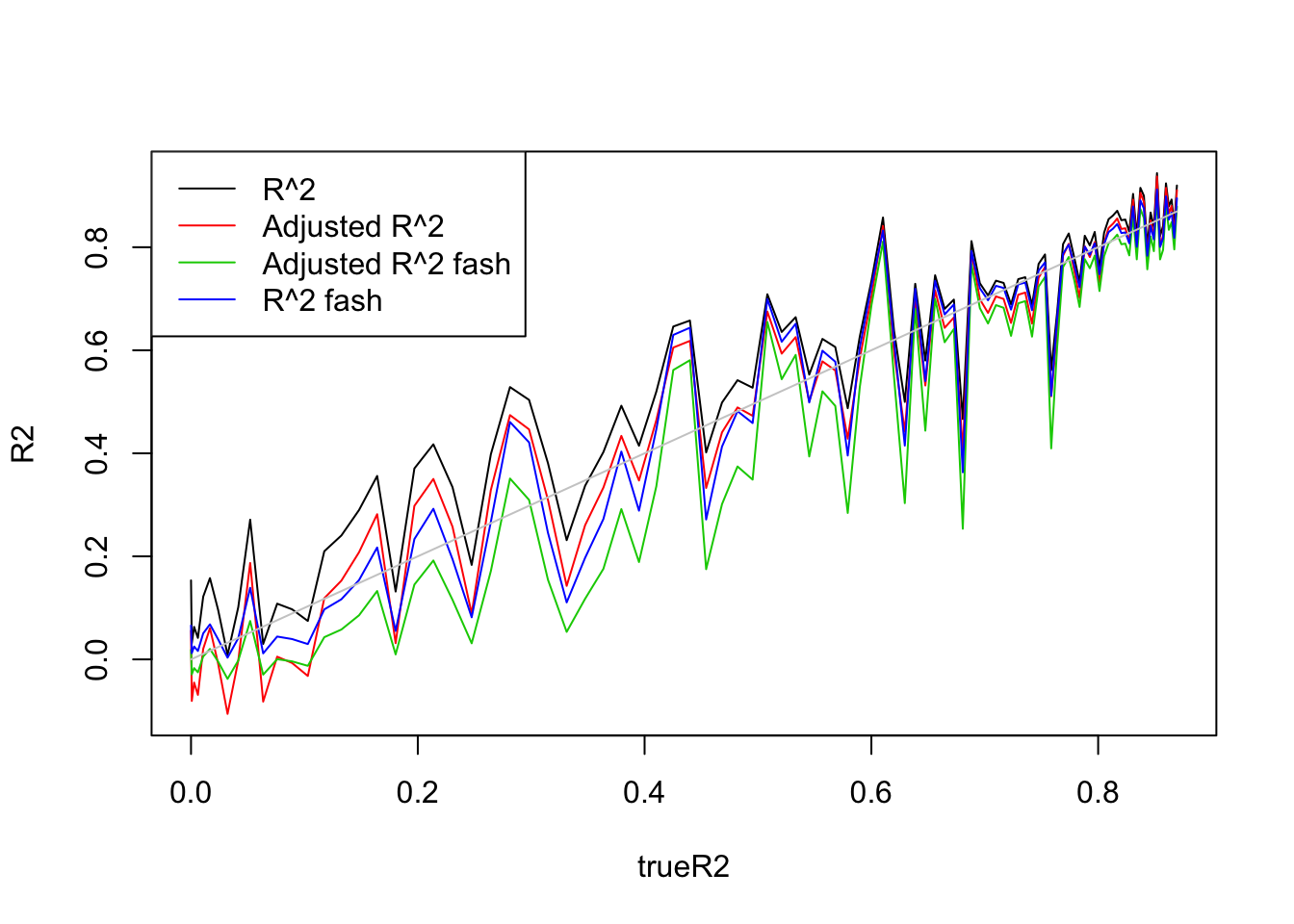
Compare fash and corshrink
1-d case
- \(n=100,p=1\)
Uniform(0,5):
library(CorShrink)
set.seed(1234)
n=100
p=1
R2=c()
R2a=c()
trueR2=c()
beta.list=seq(0,1,length.out = 100)
X=matrix(runif(n*(p),0,5),n,p)
for (i in 1:length(beta.list)) {
beta=rep(beta.list[i],p)
y=X*beta+rnorm(n)
datax=data.frame(X=X,y=y)
mod=lm(y~.,datax)
mod.sy=summary(mod)
R2[i]=mod.sy$r.squared
R2a[i]=mod.sy$adj.r.squared
trueR2[i]=1-(1)/(1+var(X%*%beta))
}
R2.fash=ash_r2(R2,n,p)
R2a.fash=ash_ar2(R2,n,p)
R2.cor=(CorShrinkVector(sqrt(R2),n))^2
plot(beta.list,R2,ylim=range(c(R2,R2a,R2s,R2as,trueR2)),main='',xlab='beta',ylab='',type='l')
lines(beta.list,R2a,col=2)
#lines(beta.list,R2a.fash,col=3)
lines(beta.list,R2.fash,col=3)
lines(beta.list,R2.cor,col=4)
lines(beta.list,trueR2,col='grey80')
abline(h=0,lty=2)
legend('topleft',c('R^2','Adjusted R^2','R^2 fash','R^2 CorShrink','True R^2'),lty=c(1,1,1,1,1),col=c(1,2,3,4,'grey80'))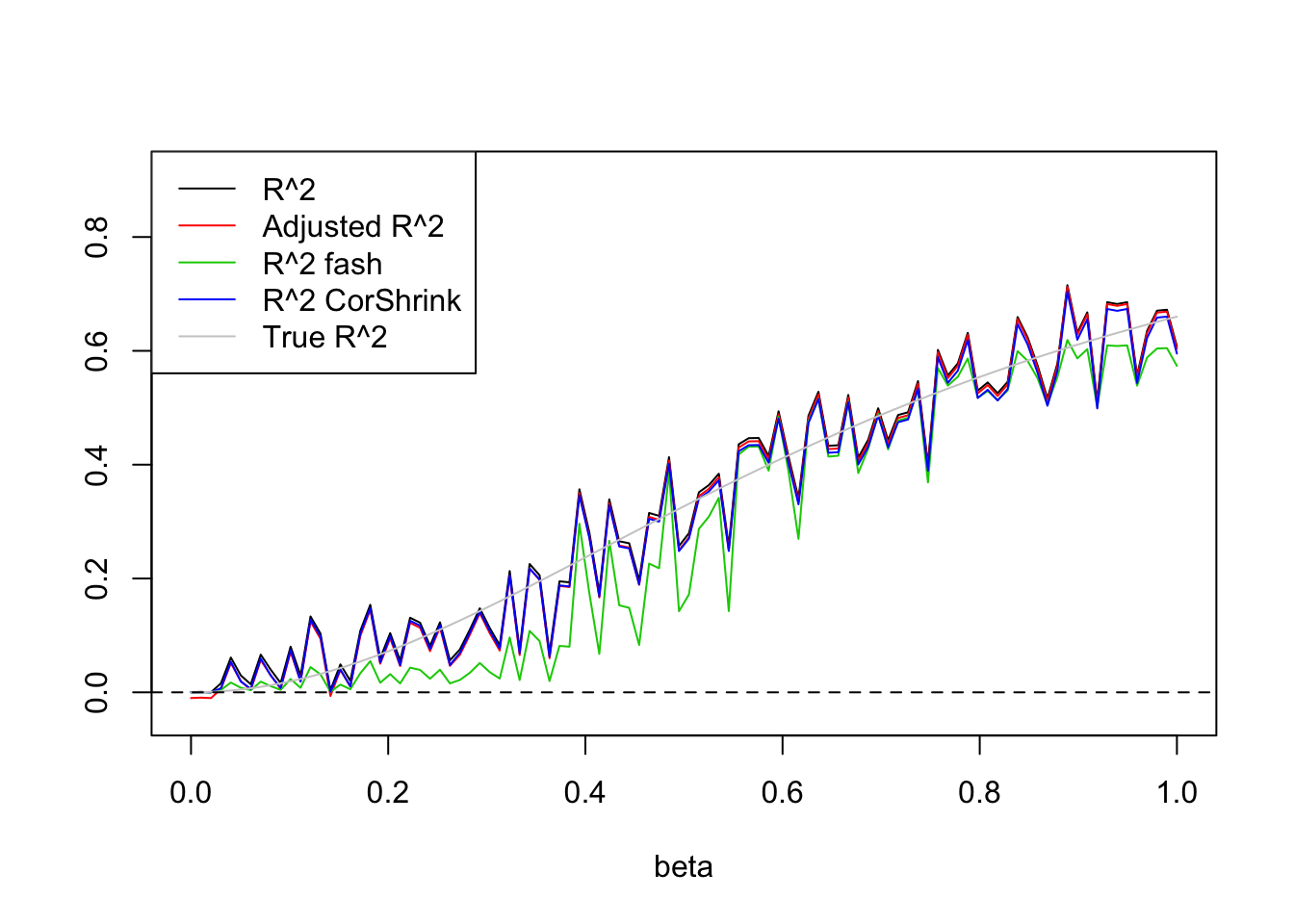
Expand here to see past versions of unnamed-chunk-7-1.png:
| Version | Author | Date |
|---|---|---|
| c6f9a91 | Dongyue Xie | 2019-02-10 |
| c7f4704 | Dongyue Xie | 2019-01-27 |
plot(trueR2,R2,type='l',ylim=range(c(R2,R2a,R2s,R2as,trueR2)))
lines(trueR2,R2a,col=2)
lines(trueR2,R2.fash,col=3)
lines(trueR2,R2.cor,col=4)
lines(trueR2,trueR2,col='grey80')
legend('topleft',c('R^2','Adjusted R^2','R^2 fash','R^2 CorShrink'),lty=c(1,1,1,1),col=c(1,2,3,4))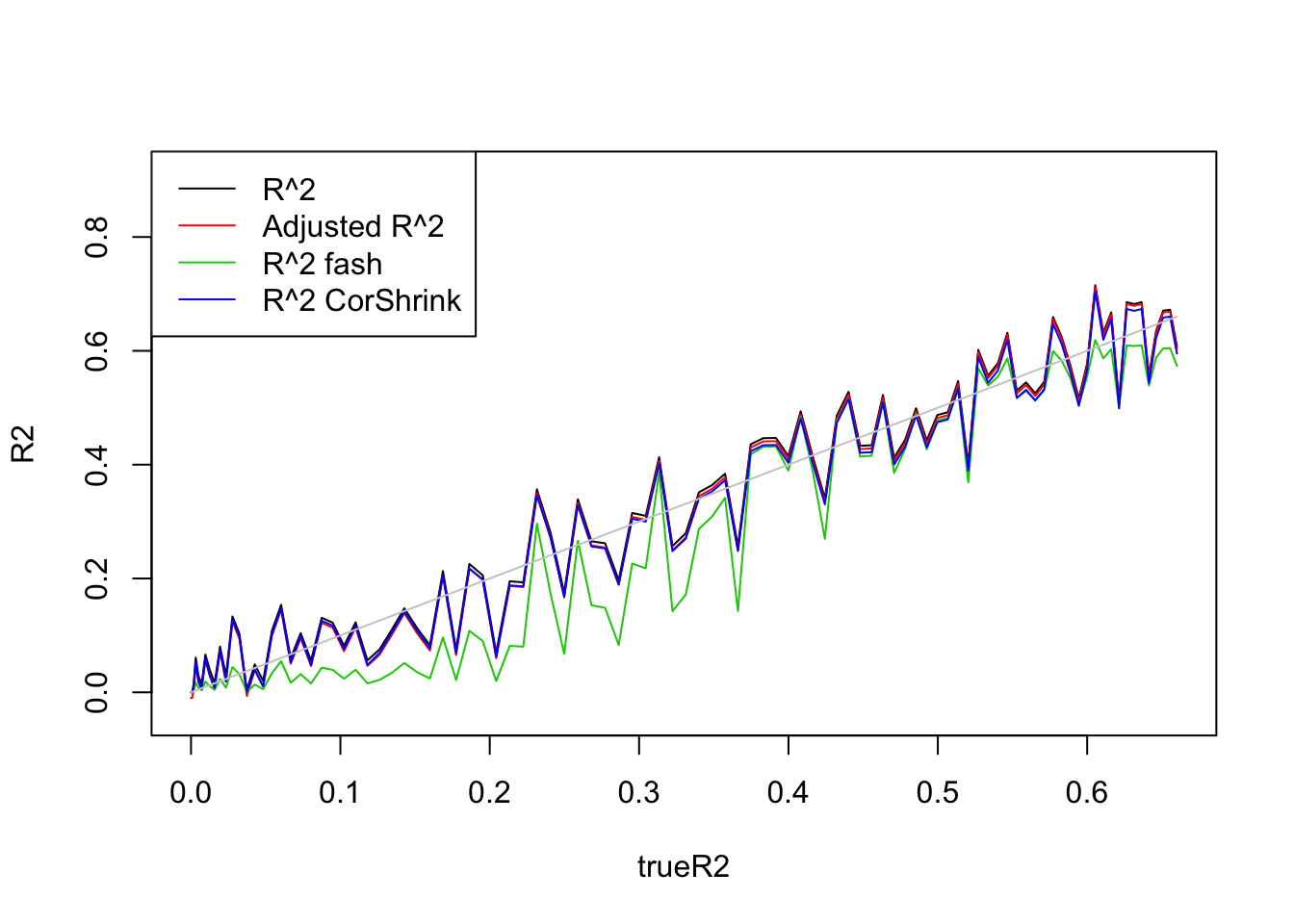
Expand here to see past versions of unnamed-chunk-7-2.png:
| Version | Author | Date |
|---|---|---|
| c6f9a91 | Dongyue Xie | 2019-02-10 |
| c7f4704 | Dongyue Xie | 2019-01-27 |
- \(n=30,p=1\)
Uniform(0,5):
set.seed(1234)
n=30
p=1
R2=c()
R2a=c()
trueR2=c()
beta.list=seq(0,1,length.out = 100)
X=matrix(runif(n*(p),0,5),n,p)
for (i in 1:length(beta.list)) {
beta=rep(beta.list[i],p)
y=X*beta+rnorm(n)
datax=data.frame(X=X,y=y)
mod=lm(y~.,datax)
mod.sy=summary(mod)
R2[i]=mod.sy$r.squared
R2a[i]=mod.sy$adj.r.squared
trueR2[i]=1-(1)/(1+var(X%*%beta))
}
R2.fash=ash_r2(R2,n,p)
R2a.fash=ash_ar2(R2,n,p)
R2.cor=(CorShrinkVector(sqrt(R2),n))^2
plot(beta.list,R2,ylim=range(c(R2,R2a,R2s,R2as,trueR2)),main='',xlab='beta',ylab='',type='l')
lines(beta.list,R2a,col=2)
#lines(beta.list,R2a.fash,col=3)
lines(beta.list,R2.fash,col=3)
lines(beta.list,R2.cor,col=4)
lines(beta.list,trueR2,col='grey80')
abline(h=0,lty=2)
legend('topleft',c('R^2','Adjusted R^2','R^2 fash','R^2 CorShrink','True R^2'),lty=c(1,1,1,1,1),col=c(1,2,3,4,'grey80'))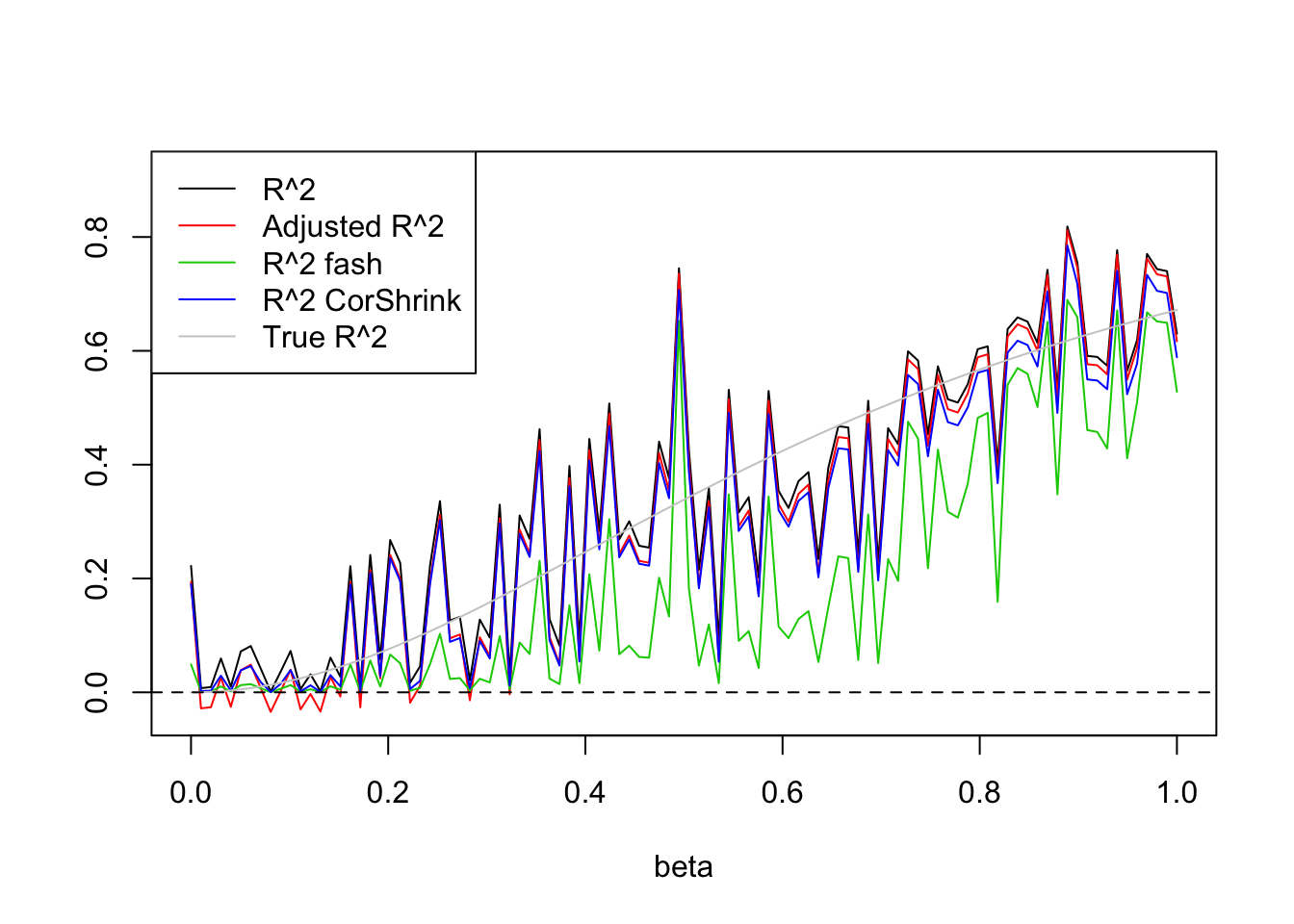
Expand here to see past versions of unnamed-chunk-8-1.png:
| Version | Author | Date |
|---|---|---|
| c6f9a91 | Dongyue Xie | 2019-02-10 |
| c7f4704 | Dongyue Xie | 2019-01-27 |
plot(trueR2,R2,type='l',ylim=range(c(R2,R2a,R2s,R2as,trueR2)))
lines(trueR2,R2a,col=2)
lines(trueR2,R2.fash,col=3)
lines(trueR2,R2.cor,col=4)
lines(trueR2,trueR2,col='grey80')
legend('topleft',c('R^2','Adjusted R^2','R^2 fash','R^2 CorShrink'),lty=c(1,1,1,1),col=c(1,2,3,4))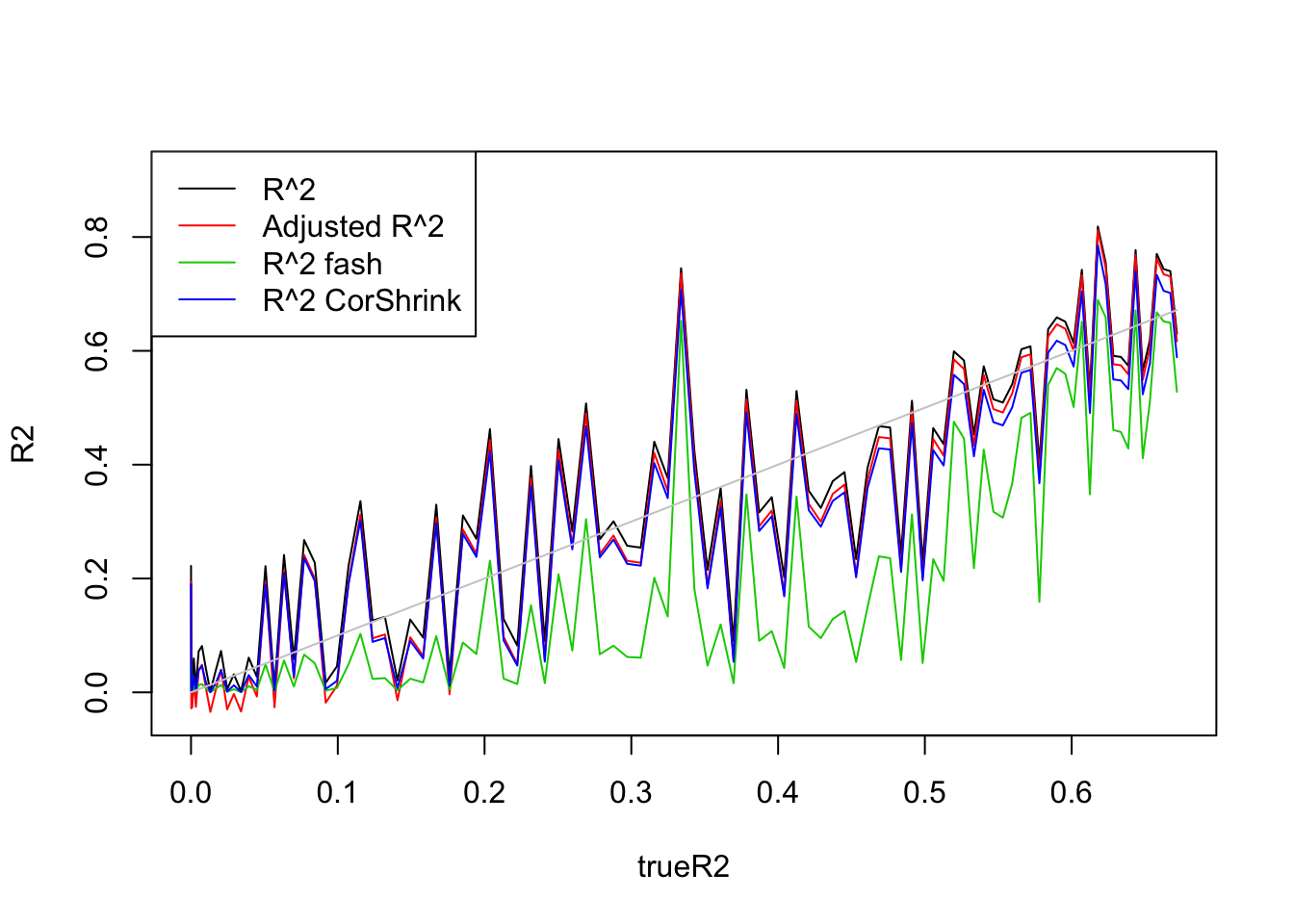
Multiple regression
- \(n=100, p=5\)
Uniform(0,2):
set.seed(1234)
n=100
p=5
R2=c()
R2a=c()
trueR2=c()
beta.list=seq(0,1,length.out = 100)
X=matrix(runif(n*(p),0,2),n,p)
for (i in 1:length(beta.list)) {
beta=rep(beta.list[i],p)
y=X%*%beta+rnorm(n)
datax=data.frame(X=X,y=y)
mod=lm(y~.,datax)
mod.sy=summary(mod)
R2[i]=mod.sy$r.squared
R2a[i]=mod.sy$adj.r.squared
trueR2[i]=1-(1)/(1+var(X%*%beta))
}
R2.fash=ash_r2(R2,n,p)
R2a.fash=ash_ar2(R2,n,p)
R2.cor=(CorShrinkVector(sqrt(R2),n))^2
plot(beta.list,R2,ylim=range(c(R2,R2a,R2s,R2as,trueR2)),main='',xlab='beta',ylab='',type='l')
lines(beta.list,R2a,col=2)
#lines(beta.list,R2a.fash,col=3)
lines(beta.list,R2.fash,col=3)
lines(beta.list,R2.cor,col=4)
lines(beta.list,trueR2,col='grey80')
abline(h=0,lty=2)
legend('topleft',c('R^2','Adjusted R^2','R^2 fash','R^2 CorShrink','True R^2'),lty=c(1,1,1,1,1),col=c(1,2,3,4,'grey80'))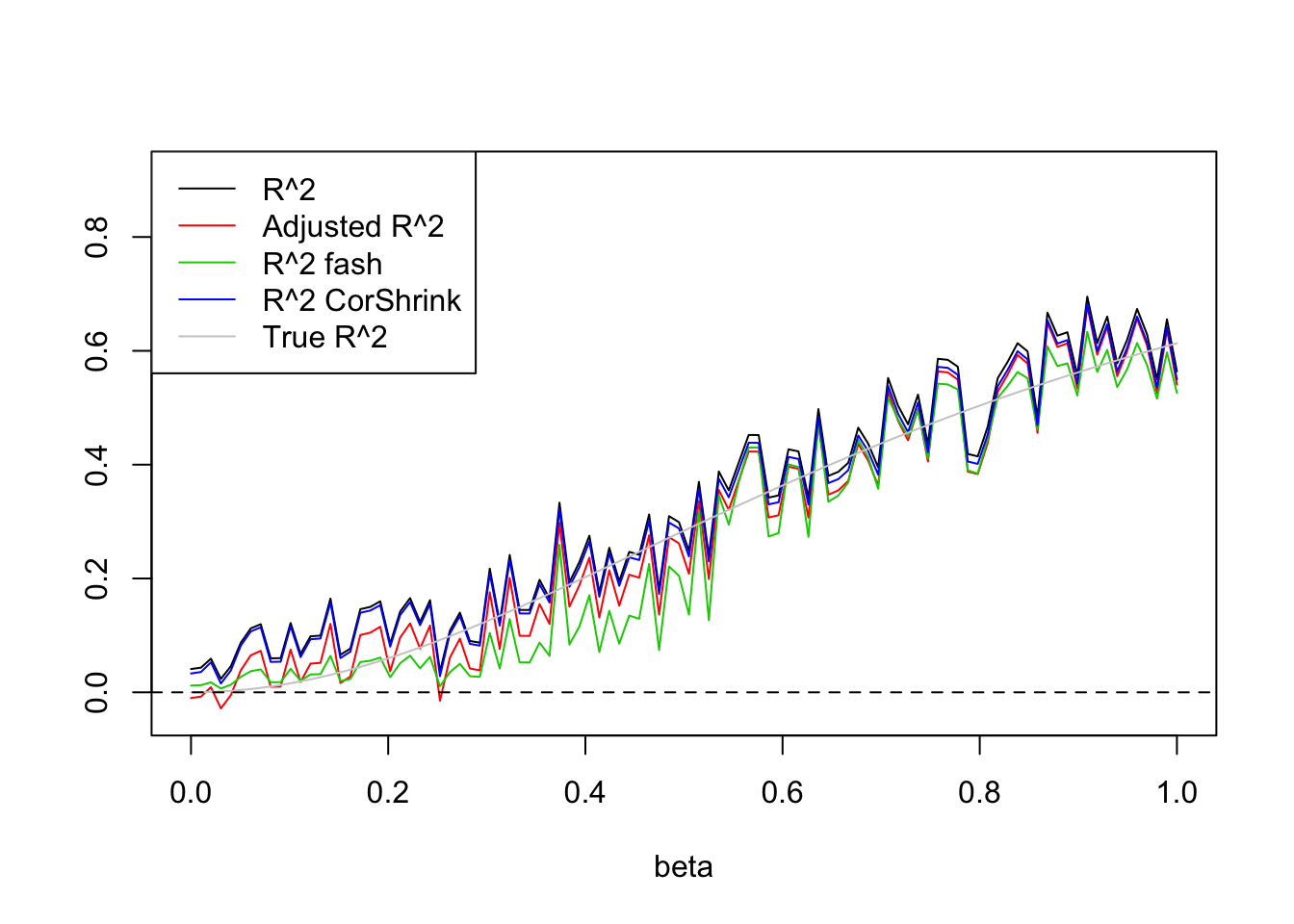
Expand here to see past versions of unnamed-chunk-9-1.png:
| Version | Author | Date |
|---|---|---|
| c6f9a91 | Dongyue Xie | 2019-02-10 |
plot(trueR2,R2,type='l',ylim=range(c(R2,R2a,R2s,R2as,trueR2)))
lines(trueR2,R2a,col=2)
lines(trueR2,R2.fash,col=3)
lines(trueR2,R2.cor,col=4)
lines(trueR2,trueR2,col='grey80')
legend('topleft',c('R^2','Adjusted R^2','R^2 fash','R^2 CorShrink'),lty=c(1,1,1,1),col=c(1,2,3,4))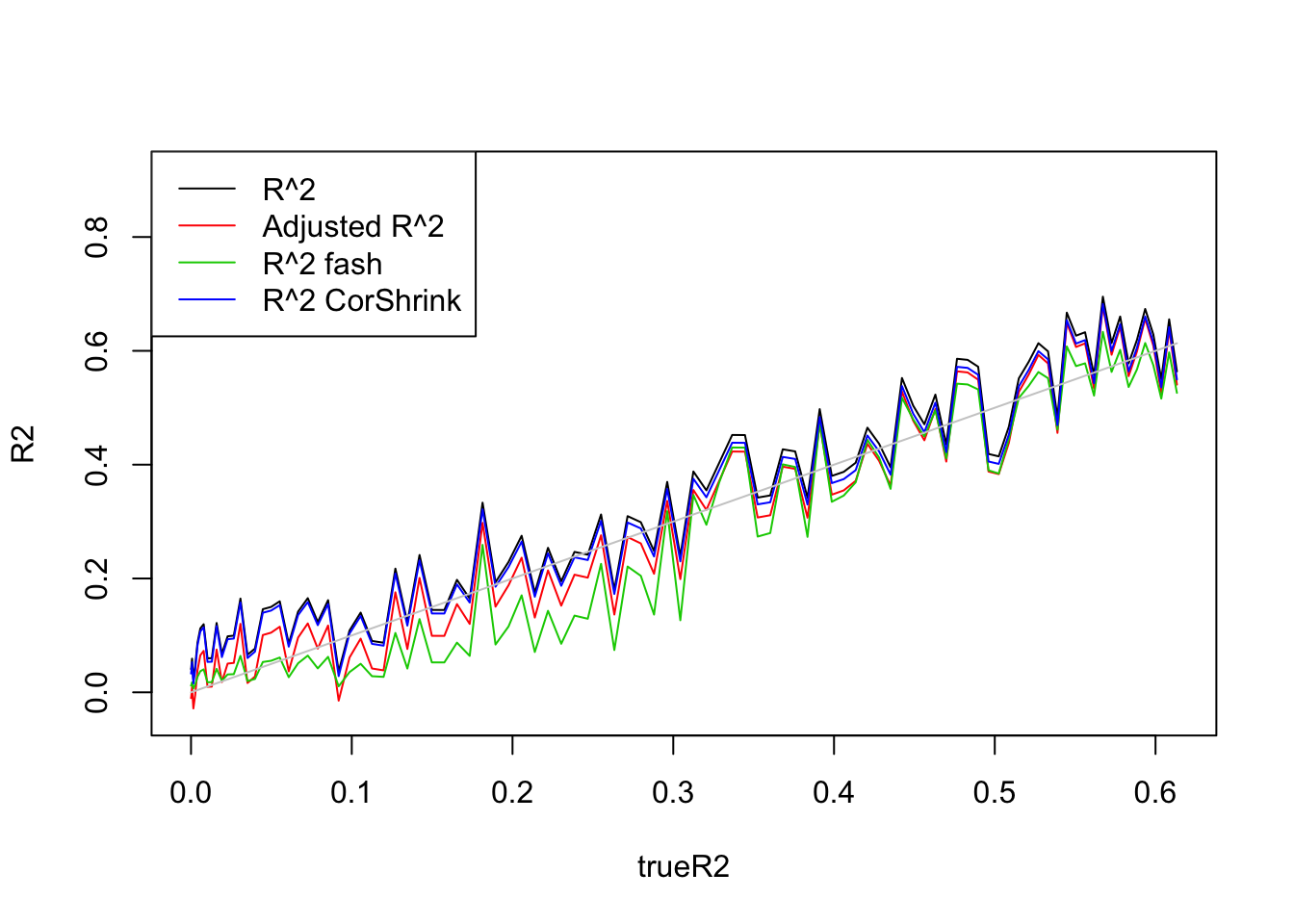
Expand here to see past versions of unnamed-chunk-9-2.png:
| Version | Author | Date |
|---|---|---|
| c6f9a91 | Dongyue Xie | 2019-02-10 |
- \(n=100,p=20\)
Uniform(0,1):
set.seed(1234)
n=100
p=20
R2=c()
R2a=c()
trueR2=c()
beta.list=seq(0,1,length.out = 100)
X=matrix(runif(n*(p),0,1),n,p)
for (i in 1:length(beta.list)) {
beta=rep(beta.list[i],p)
y=X%*%beta+rnorm(n)
datax=data.frame(X=X,y=y)
mod=lm(y~.,datax)
mod.sy=summary(mod)
R2[i]=mod.sy$r.squared
R2a[i]=mod.sy$adj.r.squared
trueR2[i]=1-(1)/(1+var(X%*%beta))
}
R2.fash=ash_r2(R2,n,p)
R2a.fash=ash_ar2(R2,n,p)
R2.cor=(CorShrinkVector(sqrt(R2),n))^2
plot(beta.list,R2,ylim=range(c(R2,R2a,R2s,R2as,trueR2)),main='',xlab='beta',ylab='',type='l')
lines(beta.list,R2a,col=2)
#lines(beta.list,R2a.fash,col=3)
lines(beta.list,R2.fash,col=3)
lines(beta.list,R2.cor,col=4)
lines(beta.list,trueR2,col='grey80')
abline(h=0,lty=2)
legend('topleft',c('R^2','Adjusted R^2','R^2 fash','R^2 CorShrink','True R^2'),lty=c(1,1,1,1,1),col=c(1,2,3,4,'grey80'))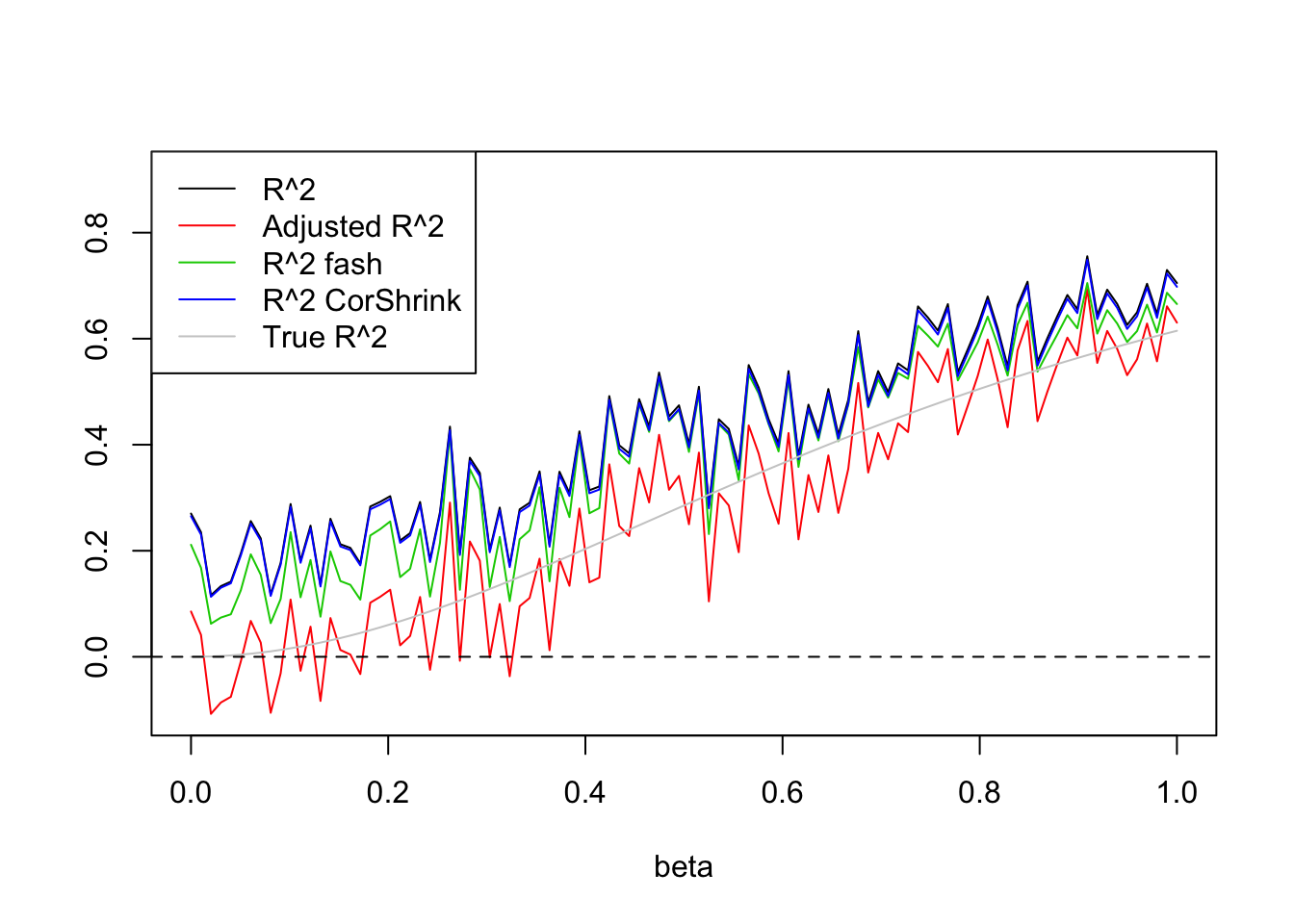
Expand here to see past versions of unnamed-chunk-10-1.png:
| Version | Author | Date |
|---|---|---|
| c6f9a91 | Dongyue Xie | 2019-02-10 |
plot(trueR2,R2,type='l',ylim=range(c(R2,R2a,R2s,R2as,trueR2)))
lines(trueR2,R2a,col=2)
lines(trueR2,R2.fash,col=3)
lines(trueR2,R2.cor,col=4)
lines(trueR2,trueR2,col='grey80')
legend('topleft',c('R^2','Adjusted R^2','R^2 fash','R^2 CorShrink'),lty=c(1,1,1,1),col=c(1,2,3,4))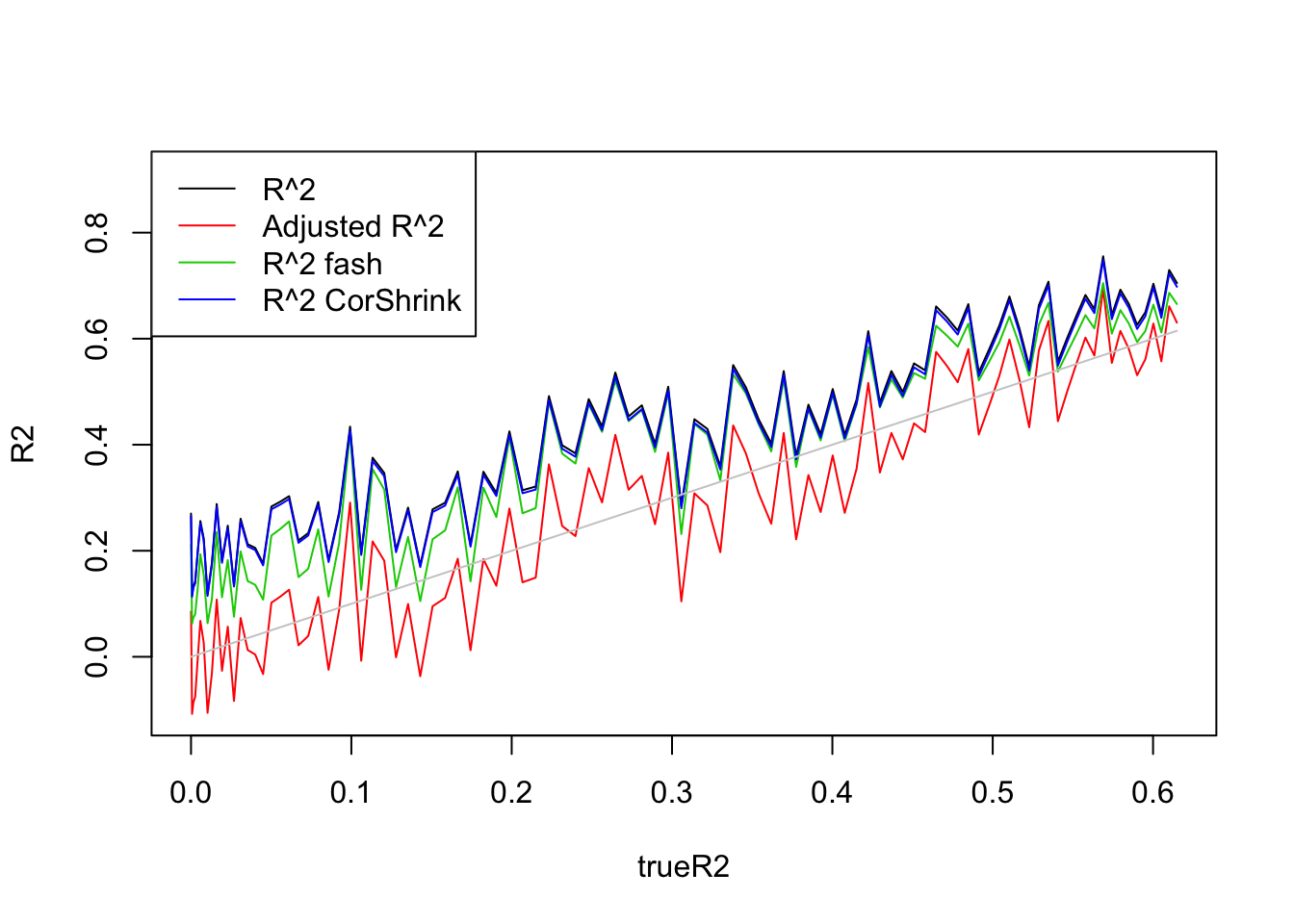
Expand here to see past versions of unnamed-chunk-10-2.png:
| Version | Author | Date |
|---|---|---|
| c6f9a91 | Dongyue Xie | 2019-02-10 |
\(n=100,p=50\)
Uniform(0,1):
set.seed(1234)
n=100
p=50
R2=c()
R2a=c()
trueR2=c()
beta.list=seq(0,1,length.out = 100)
X=matrix(runif(n*(p),0,1),n,p)
for (i in 1:length(beta.list)) {
beta=rep(beta.list[i],p)
y=X%*%beta+rnorm(n)
datax=data.frame(X=X,y=y)
mod=lm(y~.,datax)
mod.sy=summary(mod)
R2[i]=mod.sy$r.squared
R2a[i]=mod.sy$adj.r.squared
trueR2[i]=1-(1)/(1+var(X%*%beta))
}
R2.fash=ash_r2(R2,n,p)
R2a.fash=ash_ar2(R2,n,p)
R2.cor=(CorShrinkVector(sqrt(R2),n))^2
plot(beta.list,R2,ylim=range(c(R2,R2a,R2s,R2as,trueR2)),main='',xlab='beta',ylab='',type='l')
lines(beta.list,R2a,col=2)
#lines(beta.list,R2a.fash,col=3)
lines(beta.list,R2.fash,col=3)
lines(beta.list,R2.cor,col=4)
lines(beta.list,trueR2,col='grey80')
abline(h=0,lty=2)
legend('topleft',c('R^2','Adjusted R^2','R^2 fash','R^2 CorShrink','True R^2'),lty=c(1,1,1,1,1),col=c(1,2,3,4,'grey80'))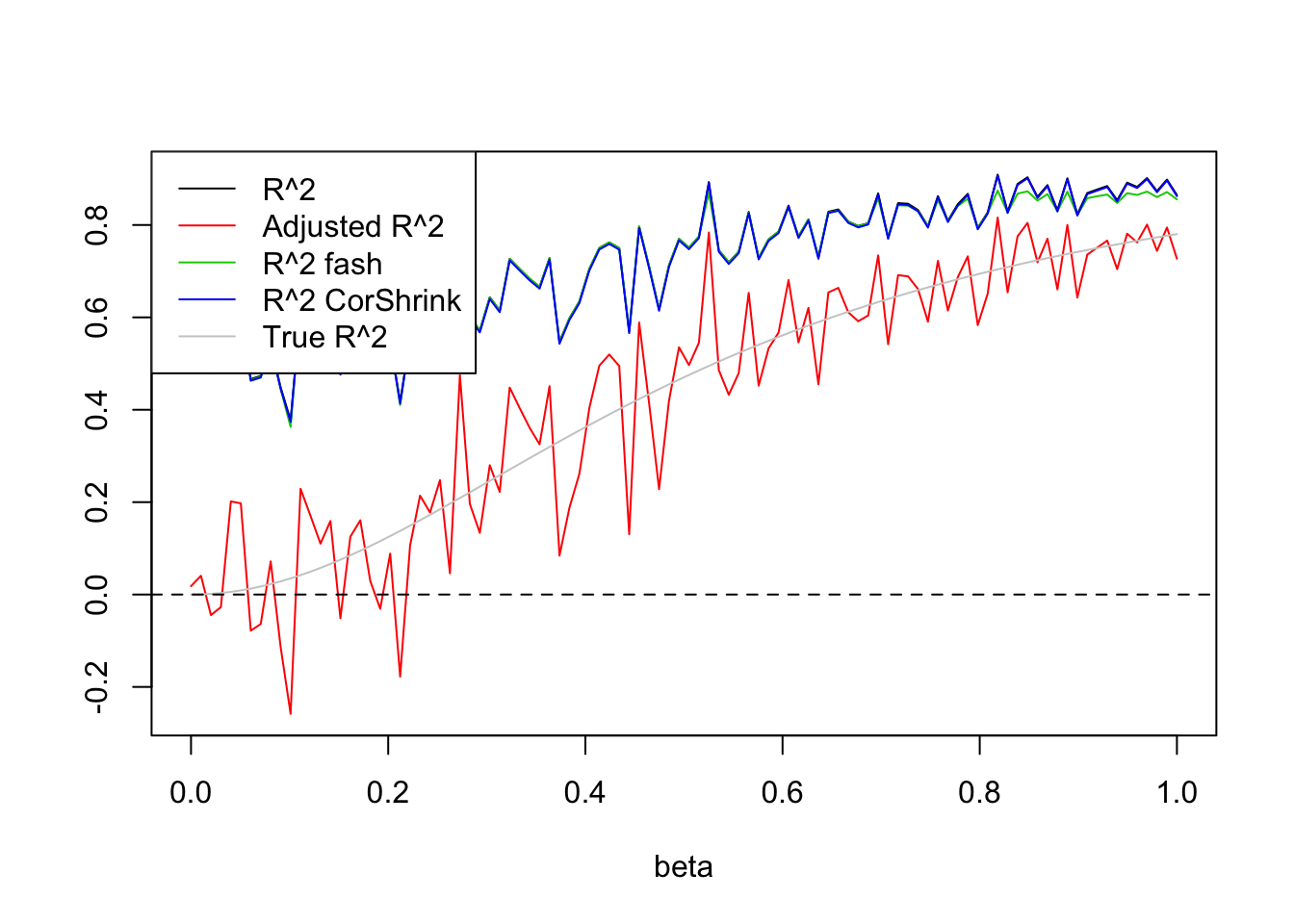
Expand here to see past versions of unnamed-chunk-11-1.png:
| Version | Author | Date |
|---|---|---|
| c6f9a91 | Dongyue Xie | 2019-02-10 |
plot(trueR2,R2,type='l',ylim=range(c(R2,R2a,R2s,R2as,trueR2)))
lines(trueR2,R2a,col=2)
lines(trueR2,R2.fash,col=3)
lines(trueR2,R2.cor,col=4)
lines(trueR2,trueR2,col='grey80')
legend('topleft',c('R^2','Adjusted R^2','R^2 fash','R^2 CorShrink'),lty=c(1,1,1,1),col=c(1,2,3,4))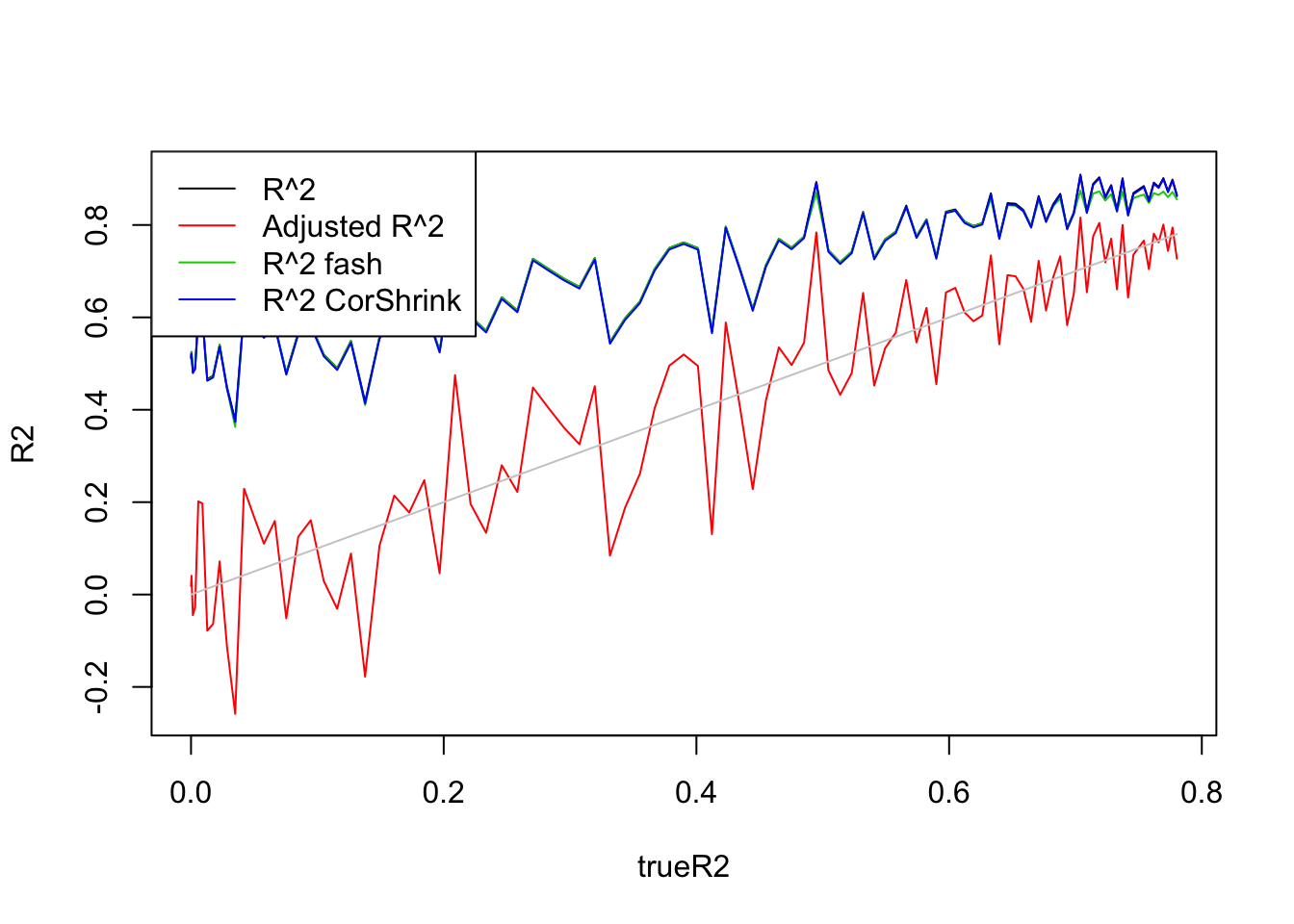
Expand here to see past versions of unnamed-chunk-11-2.png:
| Version | Author | Date |
|---|---|---|
| c6f9a91 | Dongyue Xie | 2019-02-10 |
- \(n=30, p=3\)
Uniform(0,5):
set.seed(1234)
n=30
p=3
R2=c()
R2a=c()
trueR2=c()
beta.list=seq(0,5,length.out = 100)
X=matrix(runif(n*(p),0,1),n,p)
for (i in 1:length(beta.list)) {
beta=rep(beta.list[i],p)
y=X%*%beta+rnorm(n)
datax=data.frame(X=X,y=y)
mod=lm(y~.,datax)
mod.sy=summary(mod)
R2[i]=mod.sy$r.squared
R2a[i]=mod.sy$adj.r.squared
trueR2[i]=1-(1)/(1+var(X%*%beta))
}
R2.fash=ash_r2(R2,n,p)
R2a.fash=ash_ar2(R2,n,p)
R2.cor=(CorShrinkVector(sqrt(R2),n))^2
plot(beta.list,R2,ylim=range(c(R2,R2a,R2s,R2as,trueR2)),main='',xlab='beta',ylab='',type='l')
lines(beta.list,R2a,col=2)
#lines(beta.list,R2a.fash,col=3)
lines(beta.list,R2.fash,col=3)
lines(beta.list,R2.cor,col=4)
lines(beta.list,trueR2,col='grey80')
abline(h=0,lty=2)
legend('topleft',c('R^2','Adjusted R^2','R^2 fash','R^2 CorShrink','True R^2'),lty=c(1,1,1,1,1),col=c(1,2,3,4,'grey80'))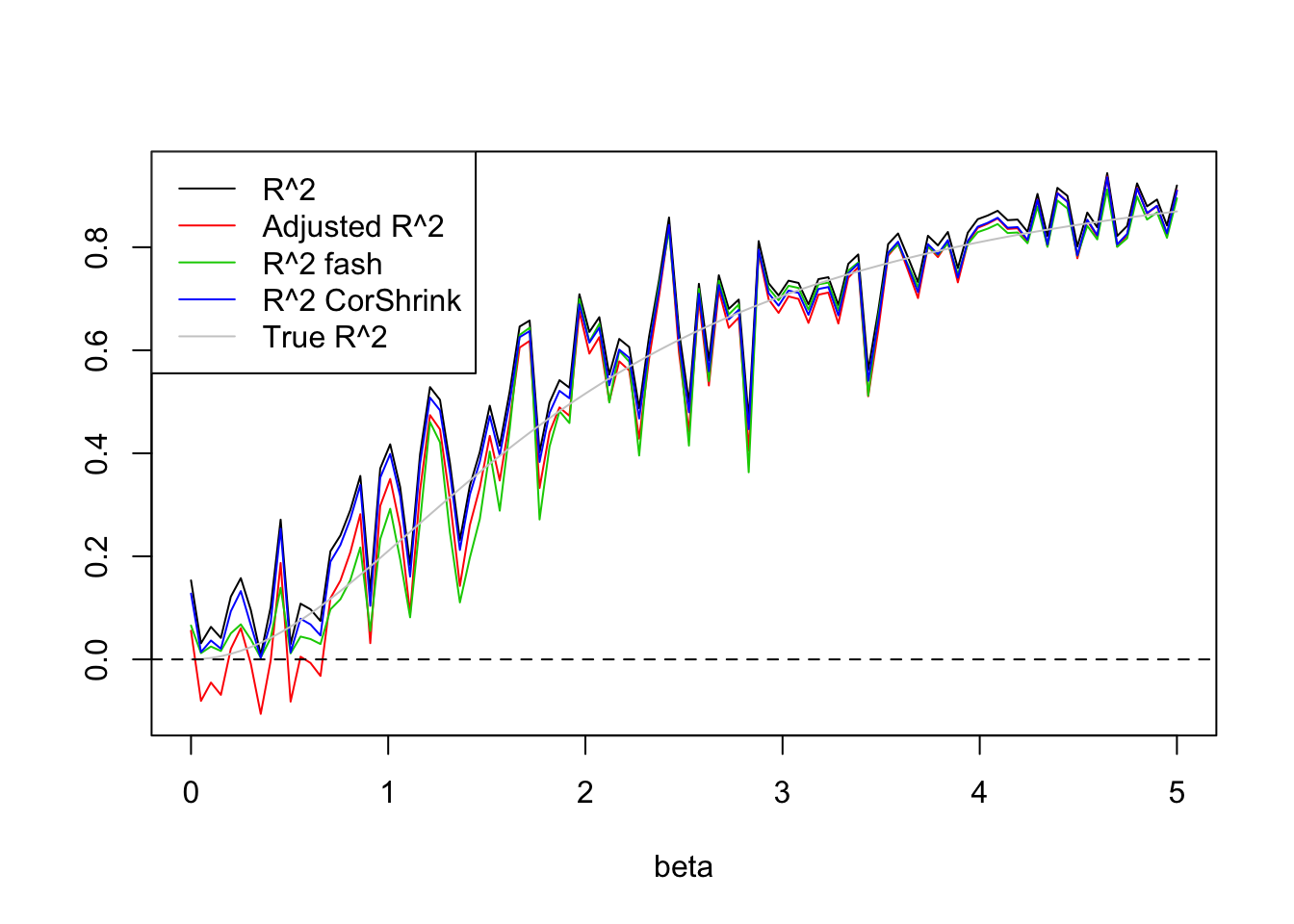
Expand here to see past versions of unnamed-chunk-12-1.png:
| Version | Author | Date |
|---|---|---|
| c6f9a91 | Dongyue Xie | 2019-02-10 |
plot(trueR2,R2,type='l',ylim=range(c(R2,R2a,R2s,R2as,trueR2)))
lines(trueR2,R2a,col=2)
lines(trueR2,R2.fash,col=3)
lines(trueR2,R2.cor,col=4)
lines(trueR2,trueR2,col='grey80')
legend('topleft',c('R^2','Adjusted R^2','R^2 fash','R^2 CorShrink'),lty=c(1,1,1,1),col=c(1,2,3,4))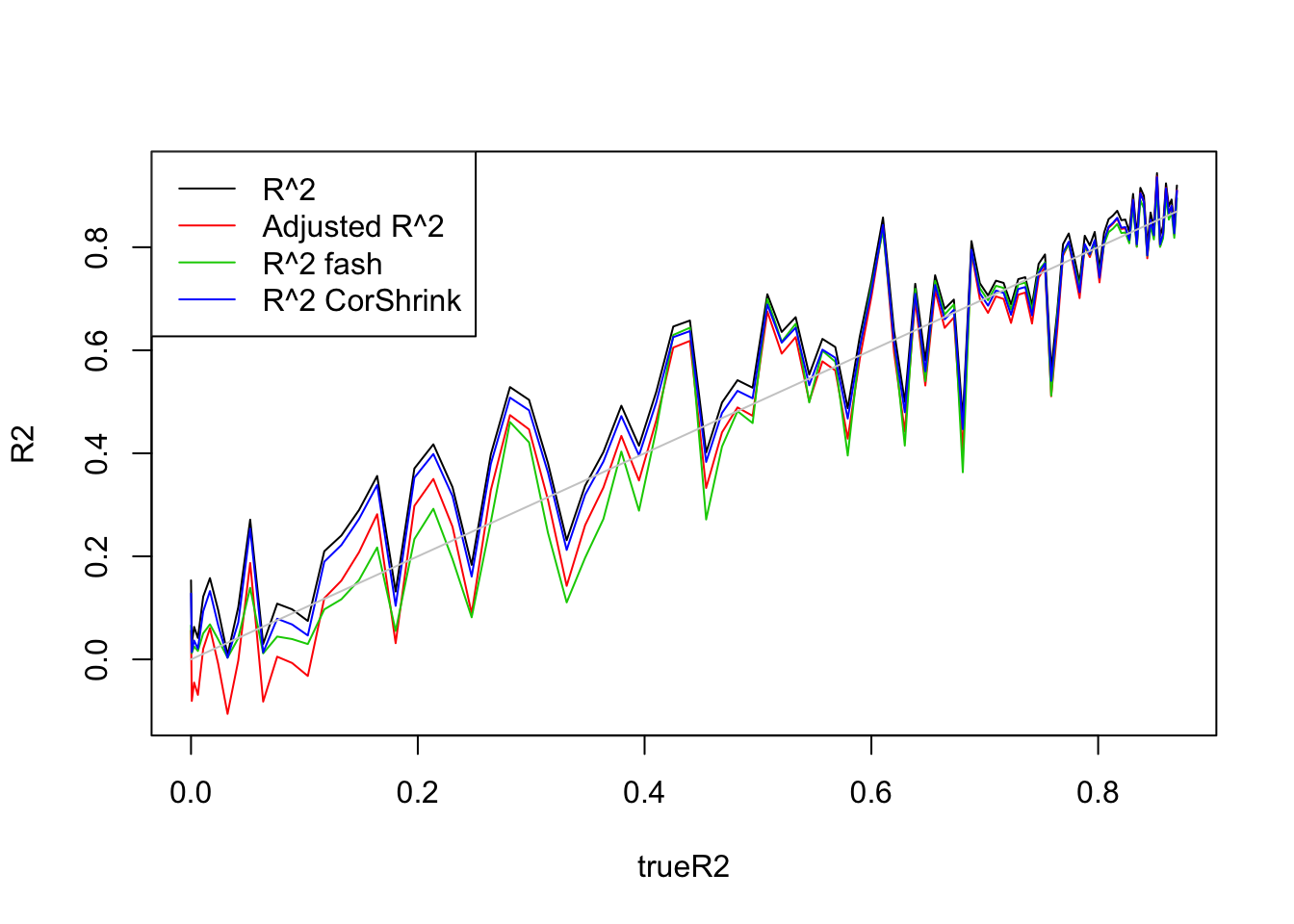
Expand here to see past versions of unnamed-chunk-12-2.png:
| Version | Author | Date |
|---|---|---|
| c6f9a91 | Dongyue Xie | 2019-02-10 |
Summary1
When generating X from Uniform(0,1), \(var(X\beta)\) is small and
fashcan shrink all \(R^2\) to 0. This happens when \(n,p\) are small. If \(p=20\), then this does not happen.CorShrink does not shrink \(R^2\). I can not really tell the difference from plots between Corshrink \(R^2\) and \(R&2\).
Adjusted \(R^2\) is a good shrinkage estimator of \(R^2\).
Sign of correlations
Randomize signs of \(R\) and see if corshrink gives the same results.
Random sample n/2 \(R^2\)s and set the sign of \(R\) to negative.
\(n=100,p=1\)
Uniform(0,1):
library(CorShrink)
set.seed(1234)
n=100
p=1
R2=c()
R2a=c()
trueR2=c()
beta.list=seq(0,1,length.out = 100)
X=matrix(runif(n*(p),0,5),n,p)
for (i in 1:length(beta.list)) {
beta=rep(beta.list[i],p)
y=X*beta+rnorm(n)
datax=data.frame(X=X,y=y)
mod=lm(y~.,datax)
mod.sy=summary(mod)
R2[i]=mod.sy$r.squared
R2a[i]=mod.sy$adj.r.squared
trueR2[i]=1-(1)/(1+var(X%*%beta))
}
R2.cor=(CorShrinkVector(sqrt(R2),n))^2
idx=sample(1:100,50)
R=sqrt(R2)
R[idx]=-R[idx]
R2.cor.sign=(CorShrinkVector(R,n))^2
plot(beta.list,R2,ylim=range(c(R2,R2.cor.sign,R2.cor,trueR2)),main='',xlab='beta',ylab='',type='l')
lines(beta.list,R2.cor,col=2)
lines(beta.list,R2.cor.sign,col=3)
lines(beta.list,trueR2,col='grey80')
abline(h=0,lty=2)
legend('topleft',c('R^2','R^2 CorShrink','R^2 CorShrink Random Sign','True R^2'),lty=c(1,1,1,1),col=c(1,2,3,'grey80'))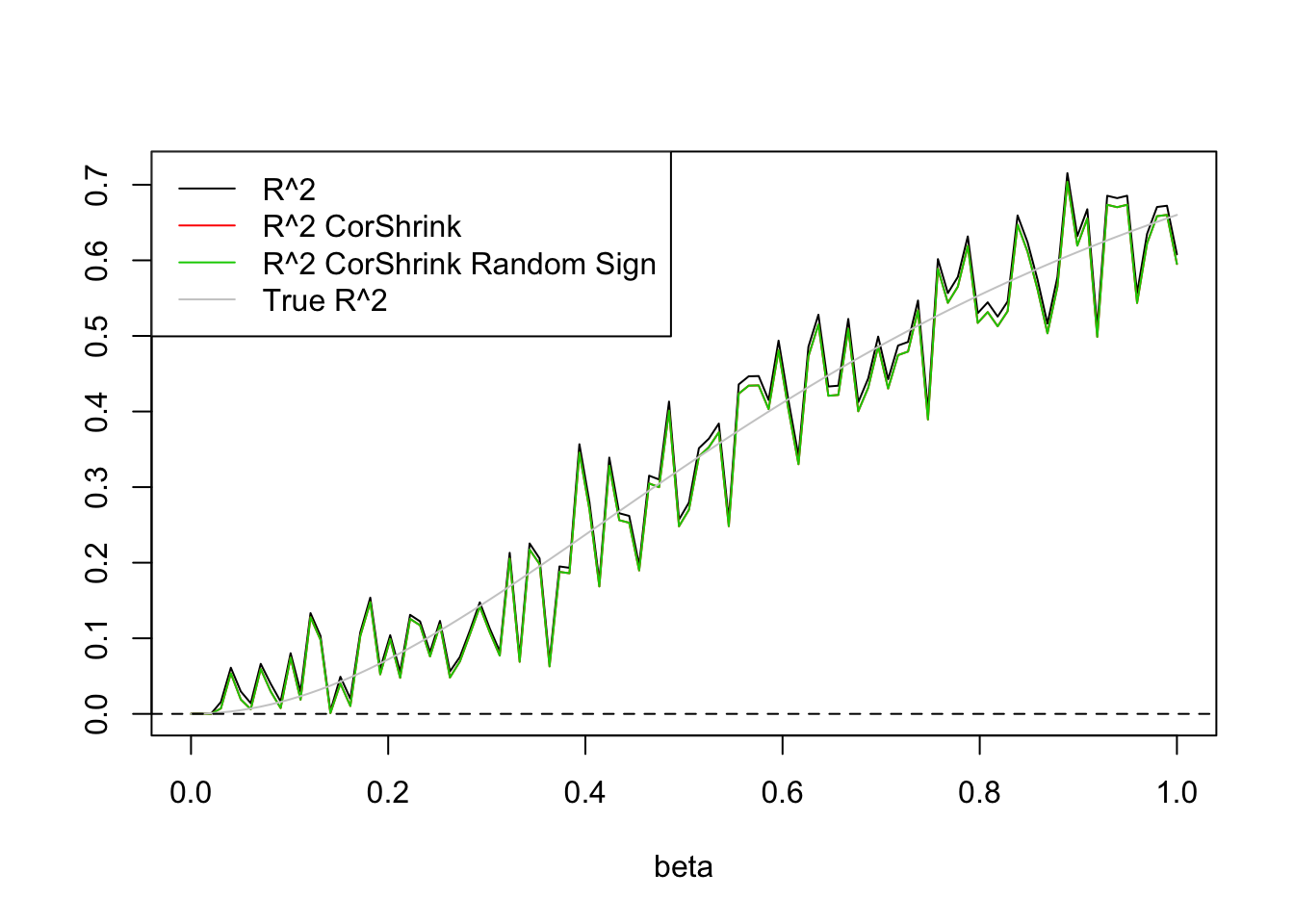
Expand here to see past versions of unnamed-chunk-13-1.png:
| Version | Author | Date |
|---|---|---|
| c6f9a91 | Dongyue Xie | 2019-02-10 |
plot(trueR2,R2,type='l',ylim=range(c(R2,R2.cor.sign,R2.cor,trueR2)))
lines(trueR2,R2.cor,col=2)
lines(trueR2,R2.cor.sign,col=3)
lines(trueR2,trueR2,col='grey80')
legend('topleft',c('R^2','R^2 CorShrink','R^2 CorShrink Random Sign'),lty=c(1,1,1),col=c(1,2,3))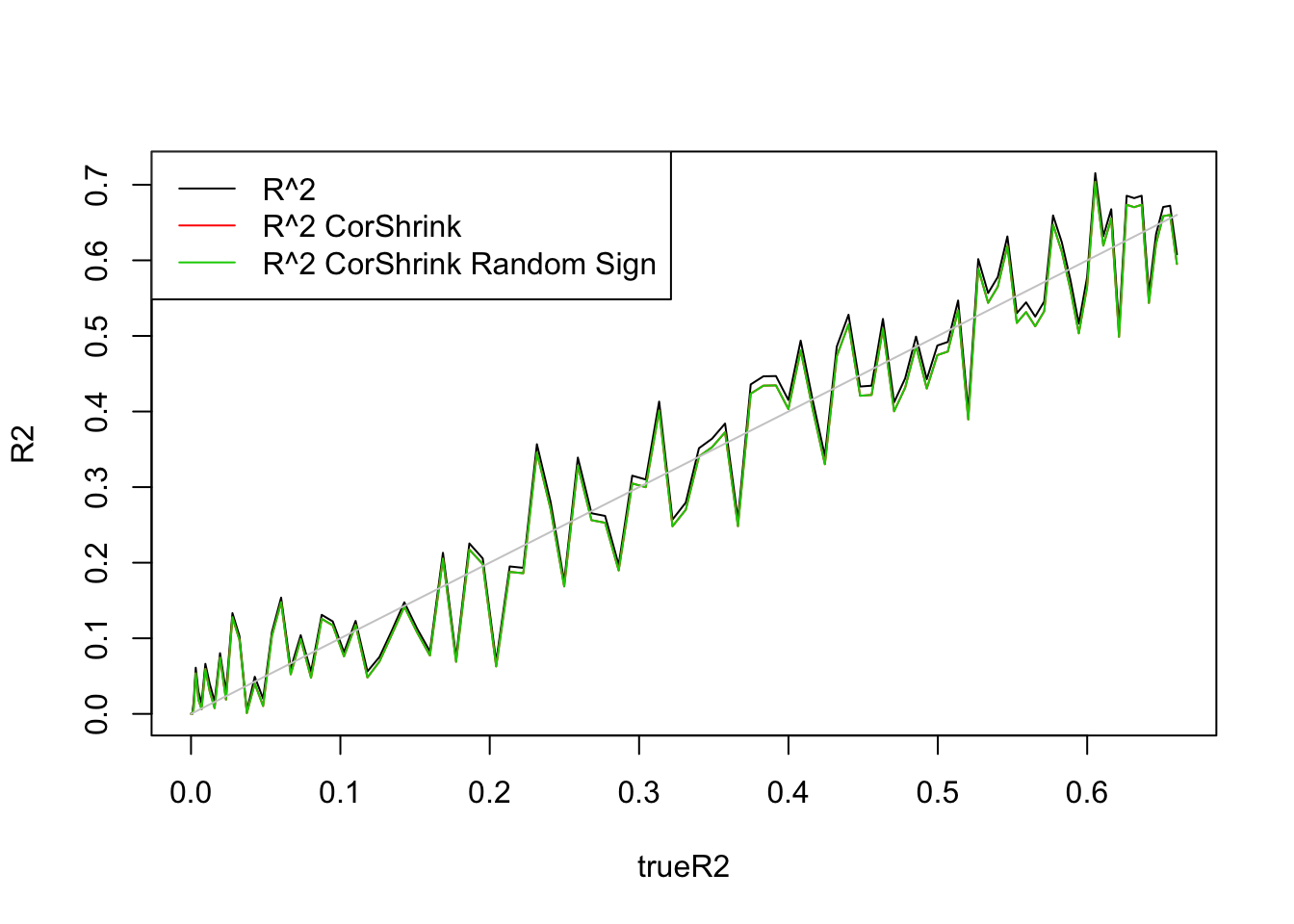
Expand here to see past versions of unnamed-chunk-13-2.png:
| Version | Author | Date |
|---|---|---|
| c6f9a91 | Dongyue Xie | 2019-02-10 |
So signs do not matter.
Estimates of g
Example 0
X from Uniform(0,5) and \(n=100,p=1\)
set.seed(1234)
n=100
p=1
R2=c()
R2a=c()
trueR2=c()
beta.list=seq(0,1,length.out = 100)
X=matrix(runif(n*(p),0,5),n,p)
for (i in 1:length(beta.list)) {
beta=rep(beta.list[i],p)
y=X%*%beta+rnorm(n)
datax=data.frame(X=X,y=y)
mod=lm(y~.,datax)
mod.sy=summary(mod)
R2[i]=mod.sy$r.squared
R2a[i]=mod.sy$adj.r.squared
trueR2[i]=1-(1)/(1+var(X%*%beta))
}
R2.fash=ash_r2(R2,n,p)
R2.cor=(CorShrinkVector(sqrt(R2),n))^2
plot(trueR2,R2.fash,type='l',ylim=range(c(R2.fash,R2.cor,trueR2)),ylab = '')
lines(trueR2,R2.cor,col=2)
lines(trueR2,trueR2,col='grey80')
legend('topleft',c('fash','Corshrink','True R2'),lty=c(1,1,1),col=c(1,2,'grey80'))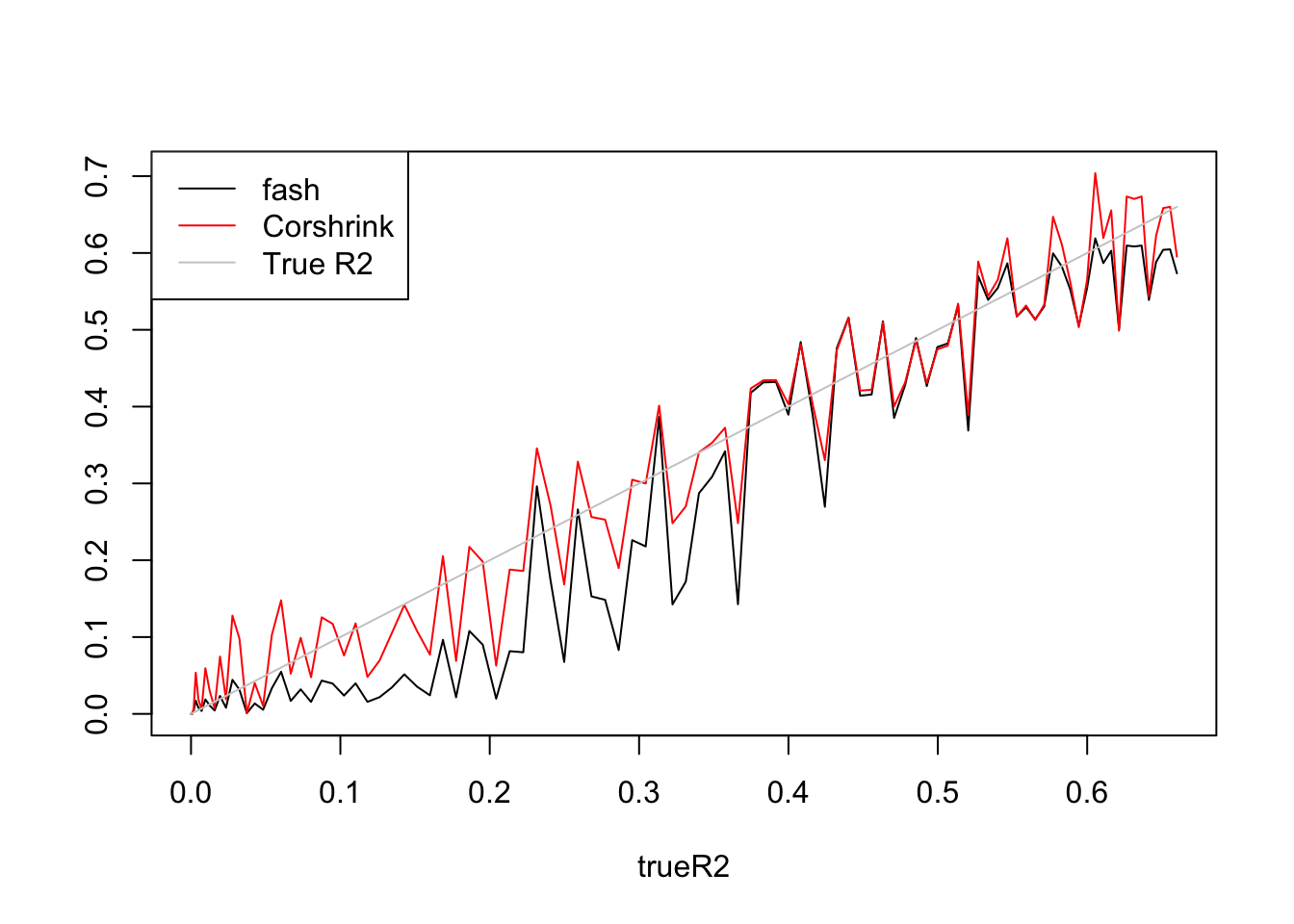
Expand here to see past versions of unnamed-chunk-14-1.png:
| Version | Author | Date |
|---|---|---|
| c6f9a91 | Dongyue Xie | 2019-02-10 |
fash:
log.ratio=log(1-R2)
ash.fit=ash(log.ratio,1,lik=lik_logF(df1=n-1,df2=n-1))
ash.fit$fitted_g$pi
[1] 3.941201e-01 1.339569e-11 1.337869e-11 1.341321e-11 1.372783e-11
[6] 1.519486e-11 2.151838e-11 6.725142e-11 6.058799e-01 1.814045e-11
$a
[1] 0.00000000 -0.09507465 -0.13445586 -0.19014930 -0.26891172
[6] -0.38029861 -0.53782345 -0.76059722 -1.07564690 -1.52119443
$b
[1] 0.00000000 0.09507465 0.13445586 0.19014930 0.26891172 0.38029861
[7] 0.53782345 0.76059722 1.07564690 1.52119443
attr(,"class")
[1] "unimix"
attr(,"row.names")
[1] 1 2 3 4 5 6 7 8 9 10Fitted g concentrates at \(0.4*\delta_0+0.6*Uniform(-1.07,1.07)\)
Corshrink:
R=sqrt(R2)
corvec=R
corvec_trans = 0.5 * log((1 + corvec)/(1 - corvec))
corvec_trans_sd = rep(sqrt(1/(n - 1) + 2/(n -
1)^2), length(corvec_trans))
ash.control=list()
ash.control.default = list(pointmass = TRUE, mixcompdist = "normal",
nullweight = 10, fixg = FALSE, mode = 0, optmethod = "mixEM",
prior = "nullbiased", gridmult = sqrt(2), outputlevel = 2,
alpha = 0, df = NULL, control = list(K = 1, method = 3,
square = TRUE, step.min0 = 1, step.max0 = 1, mstep = 4,
kr = 1, objfn.inc = 1, tol = 1e-05,
trace = FALSE))
ash.control <- utils::modifyList(ash.control.default, ash.control)
fit = do.call(ashr::ash, append(list(betahat = corvec_trans,
sebetahat = corvec_trans_sd), ash.control))
fit$fitted_g$pi
[1] 1.198336e-01 3.102514e-09 3.098655e-09 3.055578e-09 2.825173e-09
[6] 1.905128e-09 2.187591e-09 7.621847e-08 3.302451e-07 3.196176e-09
[11] 0.000000e+00 0.000000e+00 9.715355e-07 5.124079e-01 3.677565e-01
[16] 5.913347e-07 0.000000e+00 0.000000e+00
$mean
[1] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
$sd
[1] 0.000000000 0.009663507 0.013666263 0.019327015 0.027332527
[6] 0.038654030 0.054665053 0.077308060 0.109330107 0.154616120
[11] 0.218660214 0.309232240 0.437320427 0.618464480 0.874640855
[16] 1.236928959 1.749281710 2.473857919
attr(,"class")
[1] "normalmix"
attr(,"row.names")
[1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18Fitted g concentrates at \(0.12*\delta_0+0.5*N(0,0.62^2)+0.37*N(0,0.87^2)\)
Example 1
X from Uniform(0,1) and \(n=100,p=5\)
set.seed(1234)
n=100
p=5
R2=c()
R2a=c()
trueR2=c()
beta.list=seq(0,1,length.out = 100)
X=matrix(runif(n*(p),0,1),n,p)
for (i in 1:length(beta.list)) {
beta=rep(beta.list[i],p)
y=X%*%beta+rnorm(n)
datax=data.frame(X=X,y=y)
mod=lm(y~.,datax)
mod.sy=summary(mod)
R2[i]=mod.sy$r.squared
R2a[i]=mod.sy$adj.r.squared
trueR2[i]=1-(1)/(1+var(X%*%beta))
}
R2.fash=ash_r2(R2,n,p)
R2.cor=(CorShrinkVector(sqrt(R2),n))^2
plot(trueR2,R2.fash,type='l',ylim=range(c(R2.fash,R2.cor,trueR2)),ylab = '')
lines(trueR2,R2.cor,col=2)
lines(trueR2,trueR2,col='grey80')
legend('topleft',c('fash','Corshrink','True R2'),lty=c(1,1,1),col=c(1,2,'grey80'))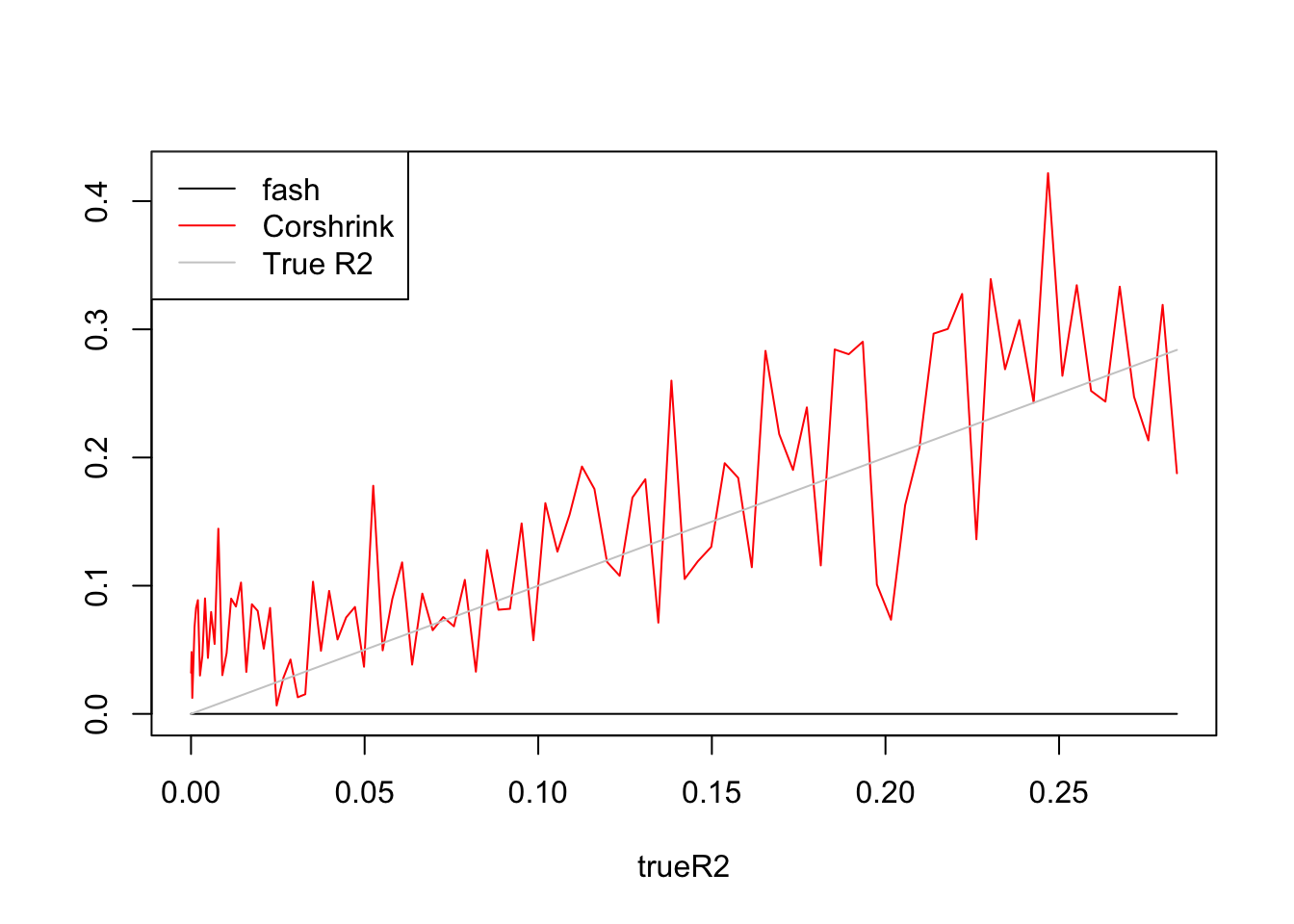
Expand here to see past versions of unnamed-chunk-17-1.png:
| Version | Author | Date |
|---|---|---|
| c6f9a91 | Dongyue Xie | 2019-02-10 |
fash:
log.ratio=log(1-R2)
ash.fit=ash(log.ratio,1,lik=lik_logF(df1=n-1,df2=n-1))
ash.fit$fitted_g$pi
[1] 1 0 0 0 0 0 0 0
$a
[1] 0.0000000 -0.1000000 -0.1414214 -0.2000000 -0.2828427 -0.4000000
[7] -0.5656854 -0.8000000
$b
[1] 0.0000000 0.1000000 0.1414214 0.2000000 0.2828427 0.4000000 0.5656854
[8] 0.8000000
attr(,"class")
[1] "unimix"
attr(,"row.names")
[1] 1 2 3 4 5 6 7 8Fitted g is at point mass at 0.
Corshrink:
R=sqrt(R2)
corvec=R
corvec_trans = 0.5 * log((1 + corvec)/(1 - corvec))
corvec_trans_sd = rep(sqrt(1/(n - 1) + 2/(n -
1)^2), length(corvec_trans))
ash.control=list()
ash.control.default = list(pointmass = TRUE, mixcompdist = "normal",
nullweight = 10, fixg = FALSE, mode = 0, optmethod = "mixEM",
prior = "nullbiased", gridmult = sqrt(2), outputlevel = 2,
alpha = 0, df = NULL, control = list(K = 1, method = 3,
square = TRUE, step.min0 = 1, step.max0 = 1, mstep = 4,
kr = 1, objfn.inc = 1, tol = 1e-05,
trace = FALSE))
ash.control <- utils::modifyList(ash.control.default, ash.control)
fit = do.call(ashr::ash, append(list(betahat = corvec_trans,
sebetahat = corvec_trans_sd), ash.control))
fit$fitted_g$pi
[1] 9.802685e-02 3.574714e-08 3.546272e-08 3.428903e-08 2.954403e-08
[6] 1.308498e-08 5.872701e-09 2.297127e-07 0.000000e+00 0.000000e+00
[11] 0.000000e+00 0.000000e+00 9.018827e-01 8.912271e-05 1.659427e-14
[16] 9.609965e-07 0.000000e+00
$mean
[1] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
$sd
[1] 0.000000000 0.009016022 0.012750581 0.018032044 0.025501162
[6] 0.036064089 0.051002323 0.072128177 0.102004647 0.144256355
[11] 0.204009293 0.288512709 0.408018586 0.577025418 0.816037173
[16] 1.154050837 1.632074345
attr(,"class")
[1] "normalmix"
attr(,"row.names")
[1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17Fitted g concentrates at \(0.9*N(0,0.41^2)\)
Example 2
X from Uniform(0,5) and \(n=100,p=5\)
set.seed(1234)
n=100
p=5
R2=c()
R2a=c()
trueR2=c()
beta.list=seq(0,1,length.out = 100)
X=matrix(runif(n*(p),0,5),n,p)
for (i in 1:length(beta.list)) {
beta=rep(beta.list[i],p)
y=X%*%beta+rnorm(n)
datax=data.frame(X=X,y=y)
mod=lm(y~.,datax)
mod.sy=summary(mod)
R2[i]=mod.sy$r.squared
R2a[i]=mod.sy$adj.r.squared
trueR2[i]=1-(1)/(1+var(X%*%beta))
}
R2.fash=ash_r2(R2,n,p)
R2.cor=(CorShrinkVector(sqrt(R2),n))^2
plot(trueR2,R2.fash,type='l',ylim=range(c(R2.fash,R2.cor,trueR2)),ylab='')
lines(trueR2,R2.cor,col=2)
lines(trueR2,trueR2,col='grey80')
legend('topleft',c('fash','Corshrink','True R2'),lty=c(1,1,1),col=c(1,2,'grey80'))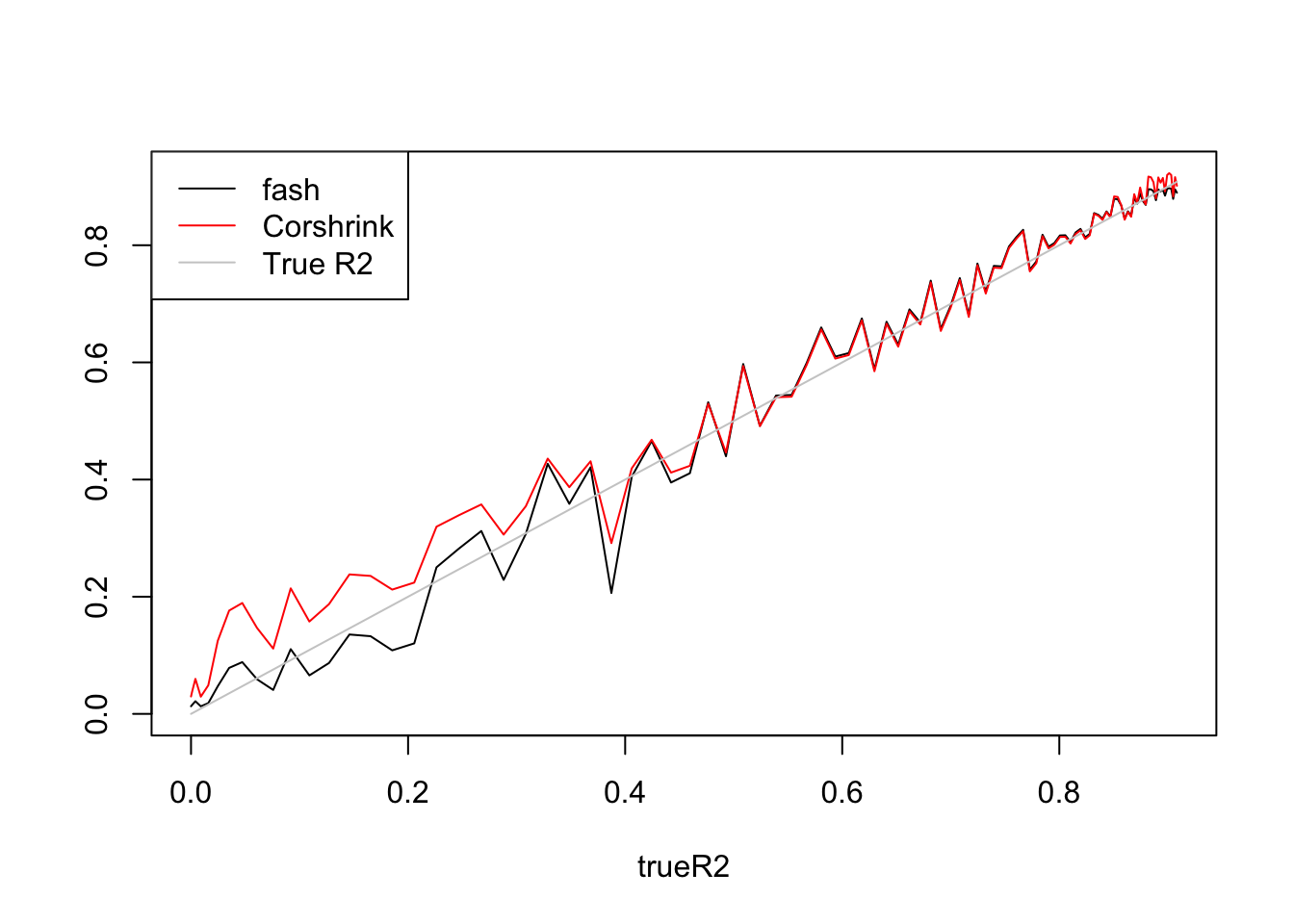
Expand here to see past versions of unnamed-chunk-20-1.png:
| Version | Author | Date |
|---|---|---|
| c6f9a91 | Dongyue Xie | 2019-02-10 |
fash:
log.ratio=log(1-R2)
ash.fit=ash(log.ratio,1,lik=lik_logF(df1=n-1,df2=n-1))
ash.fit$fitted_g$pi
[1] 1.925080e-01 3.447647e-11 3.471171e-11 3.517280e-11 3.606265e-11
[6] 3.774646e-11 4.082196e-11 4.558961e-11 5.172612e-11 6.290321e-11
[11] 1.055638e-10 8.074920e-01 4.531427e-10 4.122839e-11
$a
[1] 0.00000000 -0.07447264 -0.10532021 -0.14894527 -0.21064043
[6] -0.29789055 -0.42128085 -0.59578110 -0.84256171 -1.19156219
[11] -1.68512342 -2.38312439 -3.37024683 -4.76624878
$b
[1] 0.00000000 0.07447264 0.10532021 0.14894527 0.21064043 0.29789055
[7] 0.42128085 0.59578110 0.84256171 1.19156219 1.68512342 2.38312439
[13] 3.37024683 4.76624878
attr(,"class")
[1] "unimix"
attr(,"row.names")
[1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14Fitted g concentrates at \(0.19*\delta_0+0.81*Uniform(-2.38,2.38)\).
Corshrink:
R=sqrt(R2)
corvec=R
corvec_trans = 0.5 * log((1 + corvec)/(1 - corvec))
corvec_trans_sd = rep(sqrt(1/(n - 1) + 2/(n -
1)^2), length(corvec_trans))
ash.control=list()
ash.control.default = list(pointmass = TRUE, mixcompdist = "normal",
nullweight = 10, fixg = FALSE, mode = 0, optmethod = "mixEM",
prior = "nullbiased", gridmult = sqrt(2), outputlevel = 2,
alpha = 0, df = NULL, control = list(K = 1, method = 3,
square = TRUE, step.min0 = 1, step.max0 = 1, mstep = 4,
kr = 1, objfn.inc = 1, tol = 1e-05,
trace = FALSE))
ash.control <- utils::modifyList(ash.control.default, ash.control)
fit = do.call(ashr::ash, append(list(betahat = corvec_trans,
sebetahat = corvec_trans_sd), ash.control))
fit$fitted_g$pi
[1] 8.739018e-02 1.585320e-10 1.795248e-10 2.290505e-10 3.656616e-10
[6] 8.673704e-10 3.820458e-09 3.574858e-08 5.461574e-07 5.361614e-06
[11] 2.307459e-06 8.830531e-07 2.605247e-06 4.894297e-07 0.000000e+00
[16] 5.058173e-06 9.125923e-01 2.335972e-07 0.000000e+00 0.000000e+00
$mean
[1] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
$sd
[1] 0.000000000 0.007669231 0.010845930 0.015338461 0.021691860
[6] 0.030676923 0.043383720 0.061353845 0.086767440 0.122707690
[11] 0.173534880 0.245415381 0.347069760 0.490830762 0.694139520
[16] 0.981661524 1.388279041 1.963323048 2.776558081 3.926646095
attr(,"class")
[1] "normalmix"
attr(,"row.names")
[1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20Fitted g concentrates at \(0.91*N(0,1.39^2)\)
Summary2
Fash uses a mixture of point mass and uniform distributions as prior while CorShrink uses a mixture of point mass and normal distirbutions. Fash in these examples put more weights on point mass in fitted g than CorShrink.
Thoughts
Applying
fashto shrink \(R^2\) replies on F distribution, which is from the ratio of two variances. F distribution replies on normal assumption and independence of two normal populations. However, \(var(\sigma^2)\) and \(var(y)=var(X\beta)+var(\sigma^2)\) are not independent. So rigourously speaking, usingfashis not appropriate here.Corshirnkdepends on Fisher transformation which has bivariate normal assumption. Since \(R^2=r^2_{y,\hat y}\), we can applyCorshirnkif \(y,\hat y\) is bivariate normal distributed. By saying \(y,\hat y\) is bivariate normal distributed, I mean \(y,\hat y\) are \(n\) i.i.d samples from a bivariate normal distribution. However, this can hardly be true because \(\hat y=Hy\) where \(H=X(X^TX)^{-1}X^T\), so the \(n\) samples \(y,\hat y\) are not independently generated.From the examples above, adjusted \(R^2\) is a good estimate of true \(R^2\). It gives estimate close to ture \(R^2\) which can be seen from the True \(R^2\) - estimated \(R^2\) plot. It’s pitfall it that it’s no longer necessatily positive - it can be negative.
Session information
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS High Sierra 10.13.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] CorShrink_0.1-6 ashr_2.2-7
loaded via a namespace (and not attached):
[1] Rcpp_1.0.0 plyr_1.8.4 compiler_3.5.1
[4] git2r_0.23.0 workflowr_1.1.1 R.methodsS3_1.7.1
[7] R.utils_2.7.0 iterators_1.0.10 tools_3.5.1
[10] digest_0.6.18 corrplot_0.84 evaluate_0.11
[13] gtable_0.2.0 lattice_0.20-35 Matrix_1.2-14
[16] foreach_1.4.4 yaml_2.2.0 parallel_3.5.1
[19] gridExtra_2.3 stringr_1.3.1 knitr_1.20
[22] REBayes_1.3 rprojroot_1.3-2 grid_3.5.1
[25] glmnet_2.0-16 rmarkdown_1.10 reshape2_1.4.3
[28] corpcor_1.6.9 magrittr_1.5 whisker_0.3-2
[31] backports_1.1.2 codetools_0.2-15 htmltools_0.3.6
[34] MASS_7.3-51.1 assertthat_0.2.0 stringi_1.2.4
[37] Rmosek_8.0.69 doParallel_1.0.14 pscl_1.5.2
[40] truncnorm_1.0-8 SQUAREM_2017.10-1 R.oo_1.22.0 This reproducible R Markdown analysis was created with workflowr 1.1.1