Poisson to approximate binomial
Dongyue Xie
May 15, 2018
Last updated: 2018-05-19
workflowr checks: (Click a bullet for more information)-
✔ R Markdown file: up-to-date
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
-
✔ Environment: empty
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
-
✔ Seed:
set.seed(20180501)The command
set.seed(20180501)was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible. -
✔ Session information: recorded
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
-
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.✔ Repository version: 67830a5
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can usewflow_publishorwflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.Ignored files: Ignored: .Rhistory Ignored: .Rproj.user/ Ignored: analysis/figure/ Ignored: log/ Untracked files: Untracked: analysis/binom.Rmd Untracked: analysis/overdis.Rmd Untracked: analysis/poiwave.Rmd Untracked: analysis/smashtutorial.Rmd Untracked: data/treas_bill.csv Untracked: docs/figure/smashtutorial.Rmd/ Unstaged changes: Modified: analysis/ashpmean.Rmd Modified: analysis/nugget.Rmd
Expand here to see past versions:
Methods
Let \(X\sim Binomial(n,p)\) then \(E(X)=np, Var(X)=np(1-p)\). Poisson distribution is an approximation of binomial distribution when \(n\) is large and \(p\) is small. A rule of thumb is that \(n\geq 20, p\leq 0.05\).
Derivation: Let \(\lambda=np\)
\[\frac{n!}{x!(n-x)!}p^x(1-p)^{n-x}=\frac{n(n-1)...(n-k+1)}{x!}(\lambda/n)^x(1-\lambda/n)^{n-x}\approx \frac{\lambda^x}{x!}(1-\lambda/n)^{n-x}\] as \(n\to \infty\). Since \(lim_{n\to \infty}(1-\lambda/n)^{n}=e^{-\lambda}\) and \(lim_{n\to \infty}(1-\lambda/n)^{-x}=1\), we have \[\frac{n!}{x!(n-x)!}p^x(1-p)^{n-x}\approx \frac{\lambda^x e^{-\lambda}}{x!}.\]
If we have binomial observation \(X_t\) with \(n_t\) and treat it as Poisson observation, we can do the following expansion: \[Y_t=\log(X_t)=\log(n_tp_t)+\frac{X_t-n_tp_t}{n_tp_t}=\log(n_t)+\log(p_t)+\frac{X_t-n_tp_t}{n_tp_t}.\] This leads to \[Y_t-\log(n_t)=\log(p_t)+\frac{X_t-n_tp_t}{n_tp_t}.\]
Experiments
We compare the performance of smashgen - binomial and smashgen - poi_binom, as well as Translation Invariant (TI) thresholding (Coifman and Donoho, 1995), which is one of the best methods in a large-scale simulation study in Antoniadis et al. (2001), and Ebayesthresh (Johnstone and Silverman, 2005b).
For all experiments, T is set to be 256, nugget effect \(\sigma=1\). The mean squared errors are reported and the plots are served as visual aids.
library(smashrgen)
library(ggplot2)
library(EbayesThresh)
simu_study=function(p,sigma=1,ntri,nsimu=100,seed=12345,
niter=1,family='DaubExPhase',ashp=TRUE,verbose=FALSE,robust=FALSE,
tol=1e-2){
set.seed(seed)
smash.binom.err=c()
smash.poibinom.err=c()
ti.thresh.err=c()
eb.thresh.err=c()
n=length(p)
target=exp(p)/(1+exp(p))
for(k in 1:nsimu){
ng=rnorm(n,0,sigma)
m=exp(p+ng)
q=m/(1+m)
x=rbinom(n,ntri,q)
#fit data
smash.binom.out=smash_gen(x,dist_family = 'binomial',y_var_est='smashu',ntri=ntri)
smash.poibinom.out=smash_gen(x,dist_family = 'poi_binom',y_var_est='smashu',ntri=ntri)
ti.thresh.out=ti.thresh(x/ntri,method='rmad')
eb.thresh.out=waveti.ebayes(x/ntri)
#errors
smash.binom.err[k]=mse(target,smash.binom.out)
smash.poibinom.err[k]=mse(target,smash.poibinom.out)
ti.thresh.err[k]=mse(target,ti.thresh.out)
eb.thresh.err[k]=mse(target,eb.thresh.out)
}
return(list(est=list(smash.binom.out=smash.binom.out,smash.poibinom.out=smash.poibinom.out, ti.thresh.out=ti.thresh.out,eb.thresh.out=eb.thresh.out,x=x),
err=data.frame(smash.binom=smash.binom.err,smash.poibinom=smash.poibinom.err, ti.thresh=ti.thresh.err,eb.thresh=eb.thresh.err)))
}
waveti.ebayes = function(x, filter.number = 10, family = "DaubLeAsymm", min.level = 3) {
n = length(x)
J = log2(n)
x.w = wd(x, filter.number, family, type = "station")
for (j in min.level:(J - 1)) {
x.pm = ebayesthresh(accessD(x.w, j))
x.w = putD(x.w, j, x.pm)
}
mu.est = AvBasis(convert(x.w))
return(mu.est)
}Constant trend
\(ntri\) small, \(p\) large
The number of trials are generated from a Poisson distribution with \(\lambda=5\). \(p\) is around 0.8.
n=256
p=rep(1.5,n)
set.seed(111)
ntri=rpois(n,5)+1
result=simu_study(p,ntri=ntri)
ggplot(df2gg(result$err),aes(x=method,y=MSE))+geom_boxplot(aes(fill=method))+labs(x='')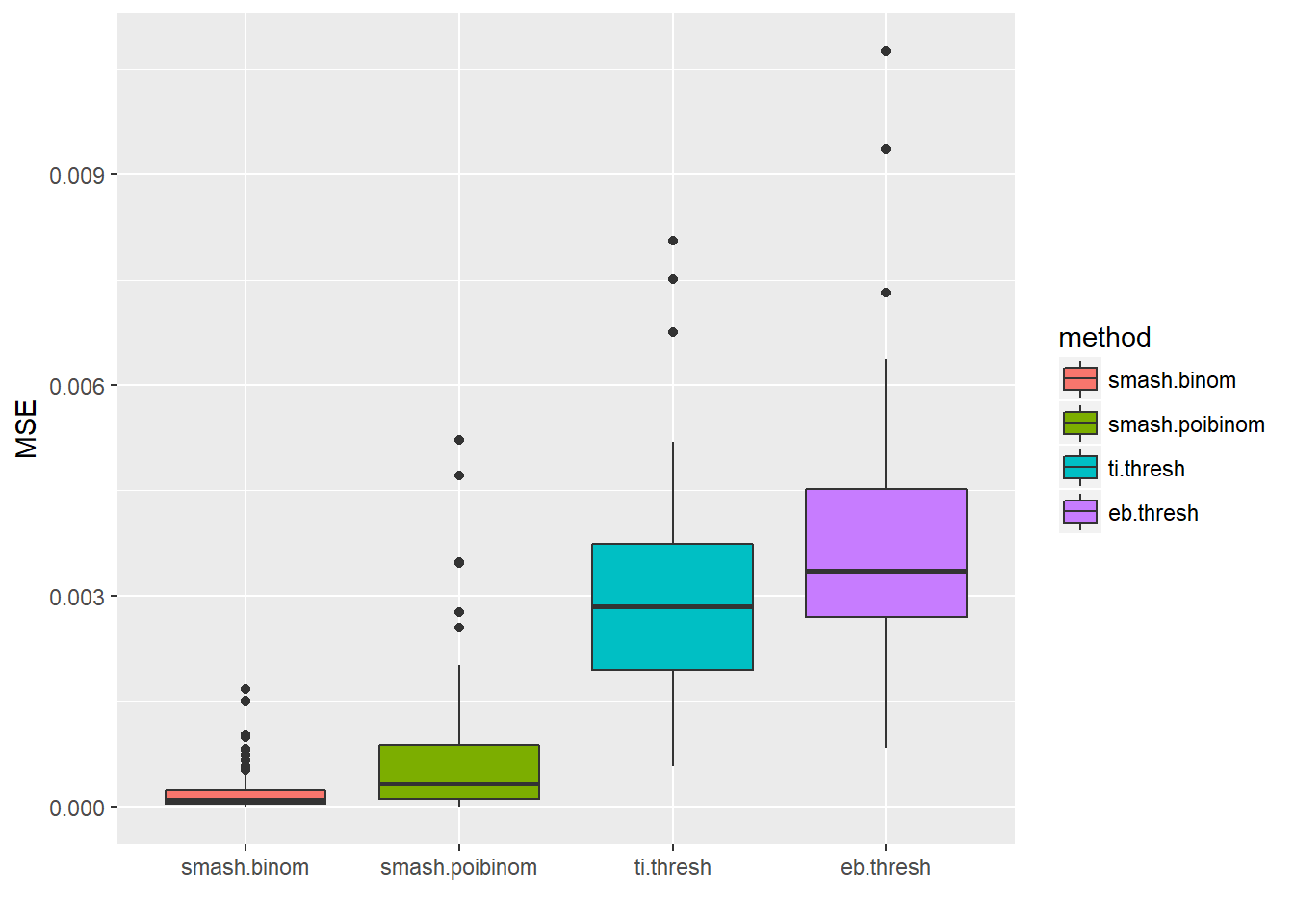
Expand here to see past versions of unnamed-chunk-2-1.png:
| Version | Author | Date |
|---|---|---|
| 9d6ca07 | Dongyue | 2018-05-18 |
apply(result$err,2,mean) smash.binom smash.poibinom ti.thresh eb.thresh
0.0002064033 0.0006906048 0.0029693084 0.0036475976 par(mfrow=c(2,2))
plot(result$est$x/ntri,col='gray80',ylab='',main='smash-binom')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$smash.binom.out,col=4,lwd=2)
legend("bottomright", # places a legend at the appropriate place
c("truth","smash-binom"), # puts text in the legend
lty=c(2,1), # gives the legend appropriate symbols (lines)
lwd=c(1,2),
cex = 1,
col=c("black","blue"))
plot(result$est$x/ntri,col='gray80',ylab='',main='smash-poi_binom')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$smash.poibinom.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='TI thresh')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$ti.thresh.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='EBayes thresh')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$eb.thresh.out,col=4,lwd=2)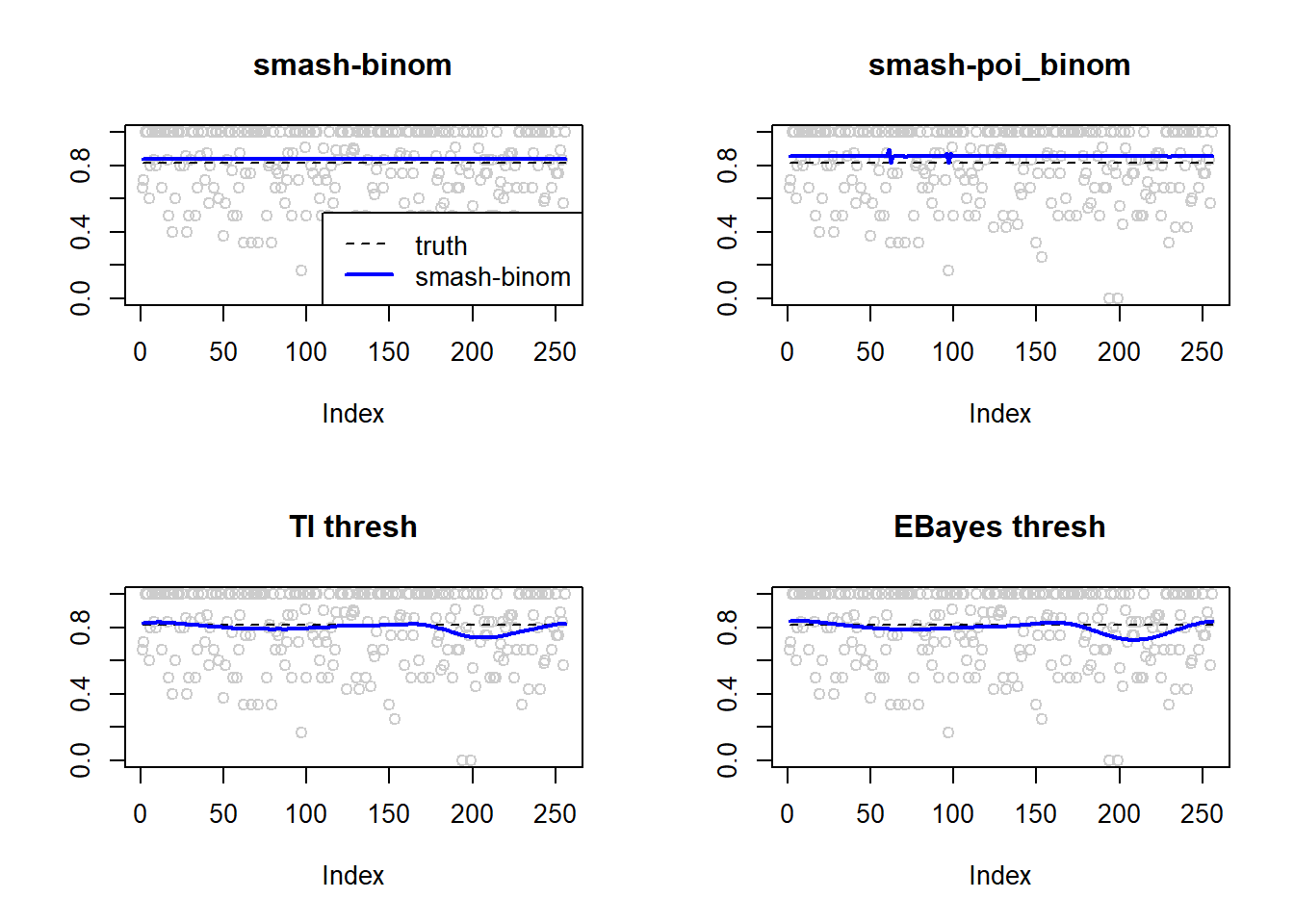
Expand here to see past versions of unnamed-chunk-2-2.png:
| Version | Author | Date |
|---|---|---|
| 9d6ca07 | Dongyue | 2018-05-18 |
\(ntri\) large, \(p\) large
We add 44 to the \(ntri\) above so that its mean is around 50.
n=256
p=rep(1.5,n)
set.seed(111)
ntri=ntri+44
result=simu_study(p,ntri=ntri)
ggplot(df2gg(result$err),aes(x=method,y=MSE))+geom_boxplot(aes(fill=method))+labs(x='')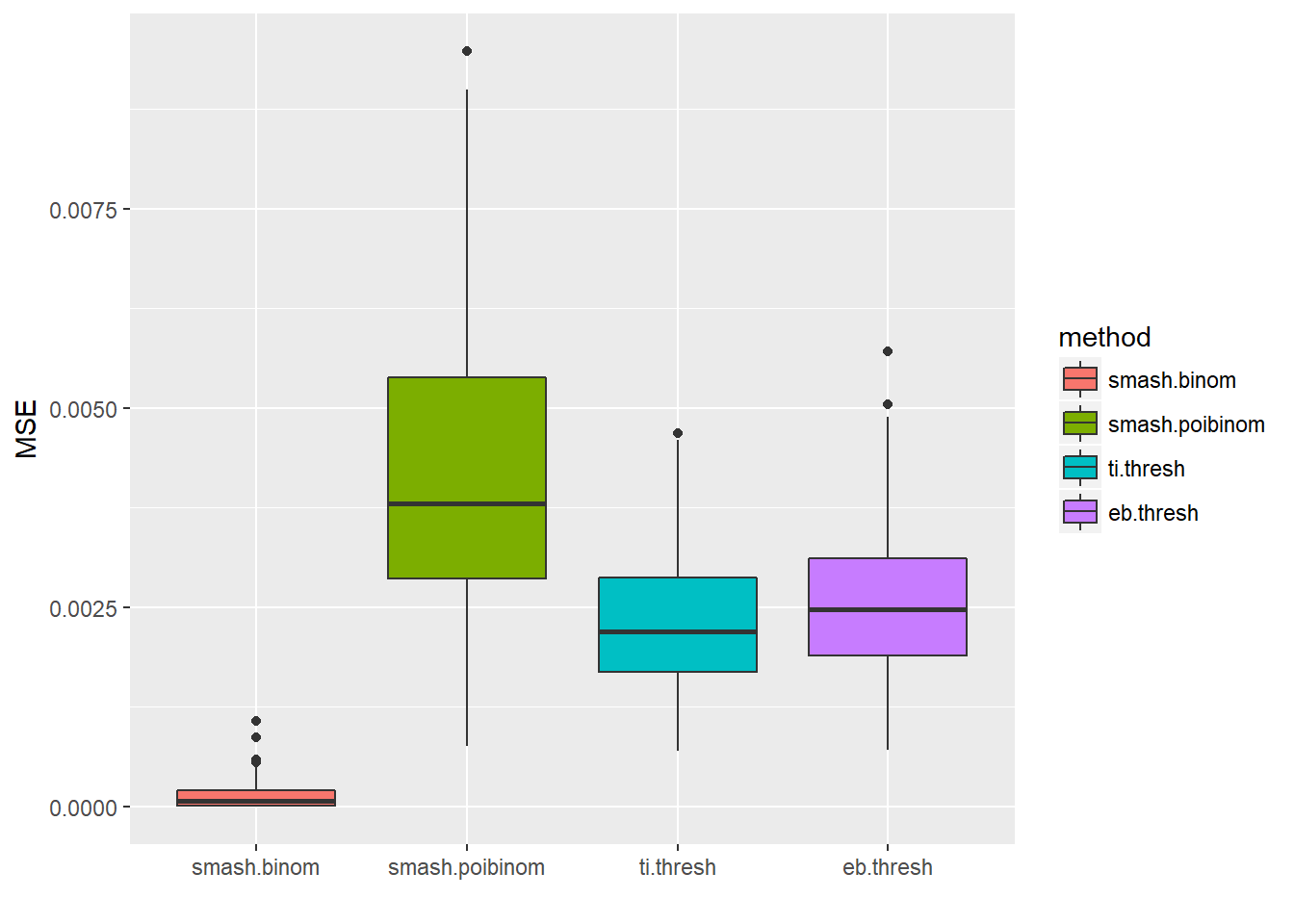
Expand here to see past versions of unnamed-chunk-3-1.png:
| Version | Author | Date |
|---|---|---|
| 9d6ca07 | Dongyue | 2018-05-18 |
apply(result$err,2,mean) smash.binom smash.poibinom ti.thresh eb.thresh
0.0001524936 0.0041117817 0.0023245279 0.0026075416 par(mfrow=c(2,2))
plot(result$est$x/ntri,col='gray80',ylab='',main='smash-binom')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$smash.binom.out,col=4,lwd=2)
legend("bottomright", # places a legend at the appropriate place
c("truth","smash-binom"), # puts text in the legend
lty=c(2,1), # gives the legend appropriate symbols (lines)
lwd=c(1,2),
cex = 1,
col=c("black","blue"))
plot(result$est$x/ntri,col='gray80',ylab='',main='smash-poi_binom')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$smash.poibinom.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='TI thresh')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$ti.thresh.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='EBayes thresh')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$eb.thresh.out,col=4,lwd=2)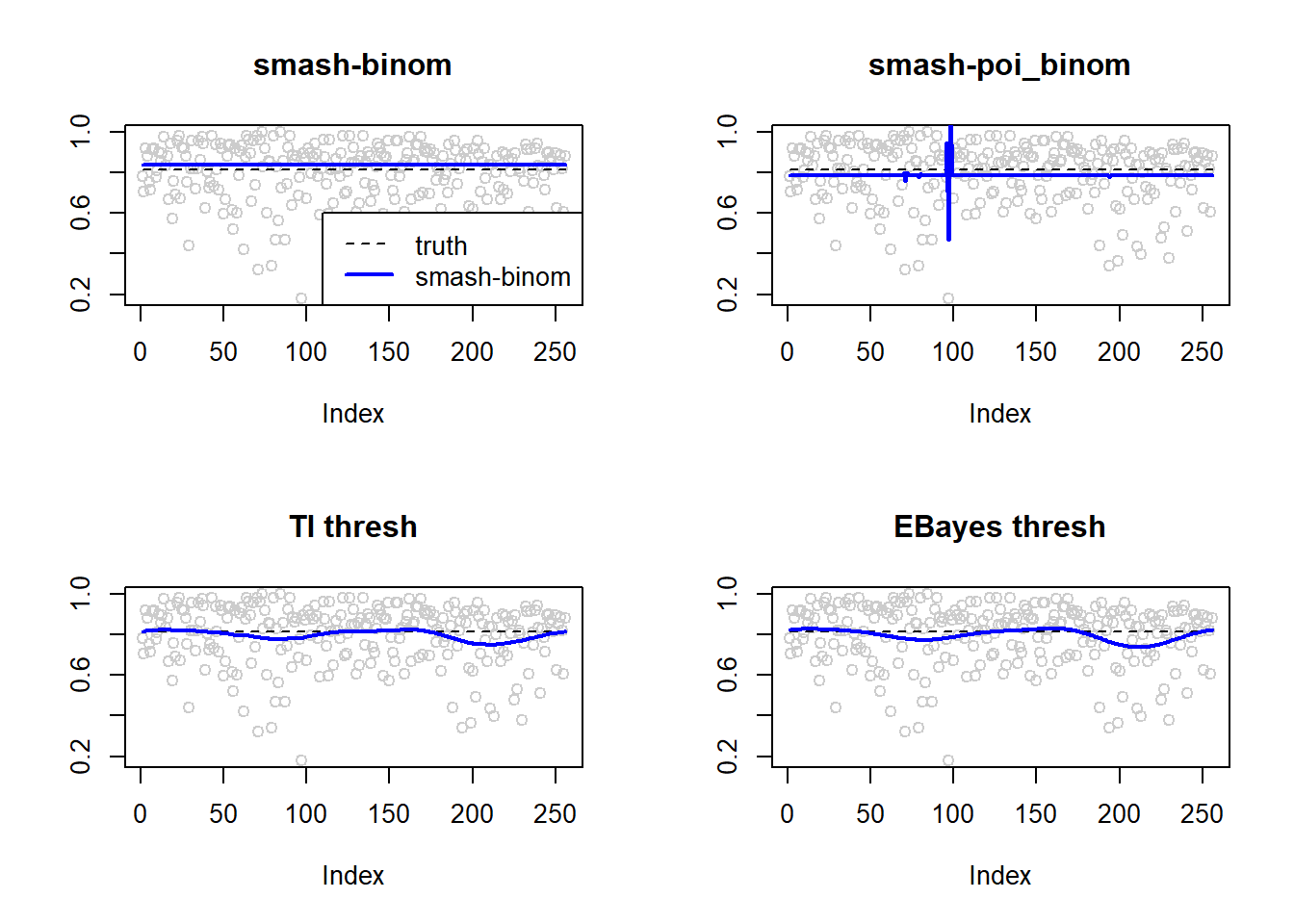
Expand here to see past versions of unnamed-chunk-3-2.png:
| Version | Author | Date |
|---|---|---|
| 9d6ca07 | Dongyue | 2018-05-18 |
\(ntri\) large, \(p\) small
\(p\) is around 0.05.
n=256
p=rep(-3,n)
set.seed(111)
ntri=ntri
result=simu_study(p,ntri=ntri)
ggplot(df2gg(result$err),aes(x=method,y=MSE))+geom_boxplot(aes(fill=method))+labs(x='')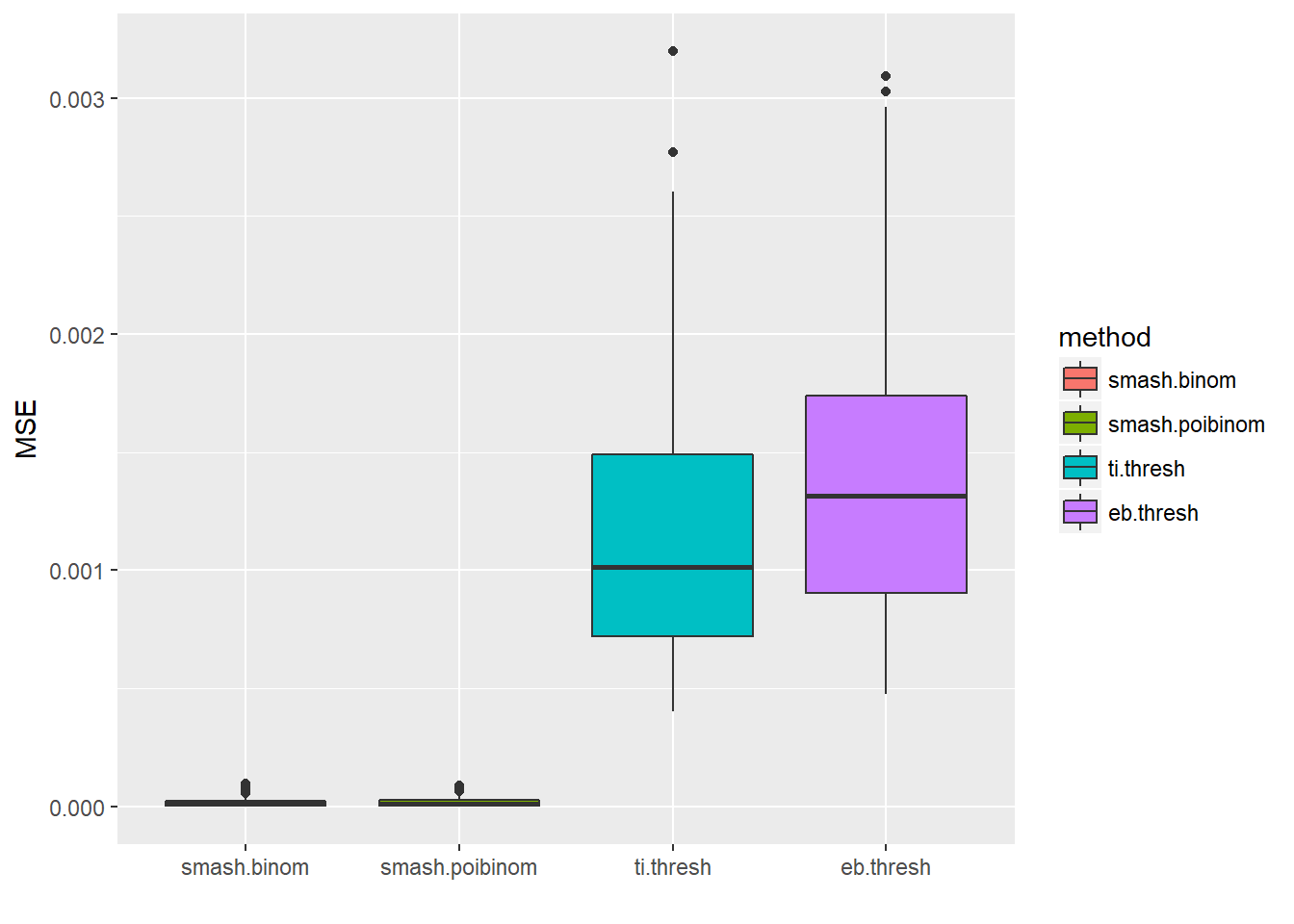
Expand here to see past versions of unnamed-chunk-4-1.png:
| Version | Author | Date |
|---|---|---|
| 9d6ca07 | Dongyue | 2018-05-18 |
ggplot(df2gg(result$err[,1:2]),aes(x=method,y=MSE))+geom_boxplot(aes(fill=method))+labs(x='')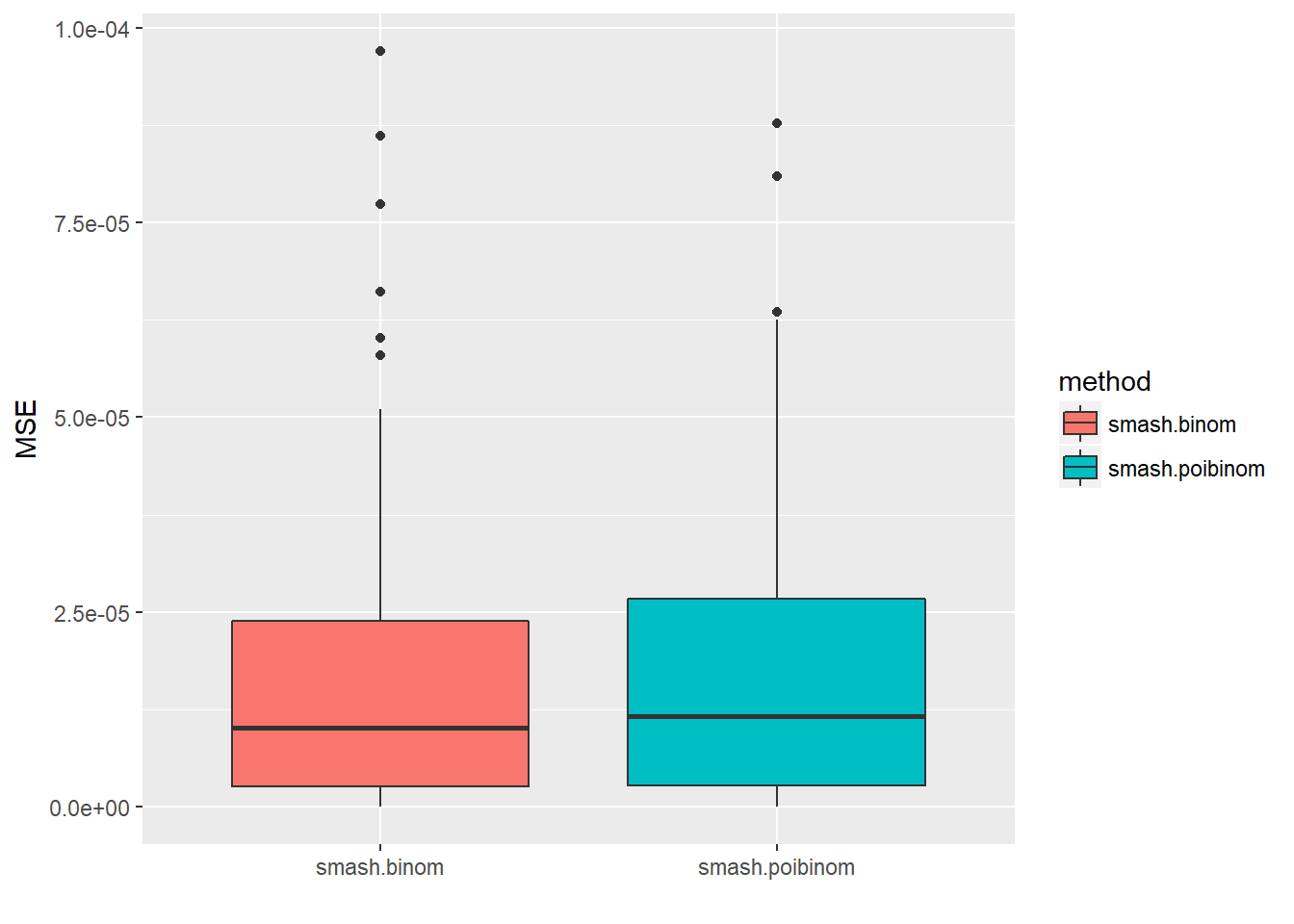
Expand here to see past versions of unnamed-chunk-4-2.png:
| Version | Author | Date |
|---|---|---|
| 9d6ca07 | Dongyue | 2018-05-18 |
apply(result$err,2,mean) smash.binom smash.poibinom ti.thresh eb.thresh
1.638404e-05 1.696551e-05 1.183316e-03 1.367054e-03 par(mfrow=c(2,2))
plot(result$est$x/ntri,col='gray80',ylab='',main='smash-binom')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$smash.binom.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='smash-poi_binom')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$smash.poibinom.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='TI thresh')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$ti.thresh.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='EBayes thresh')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$eb.thresh.out,col=4,lwd=2)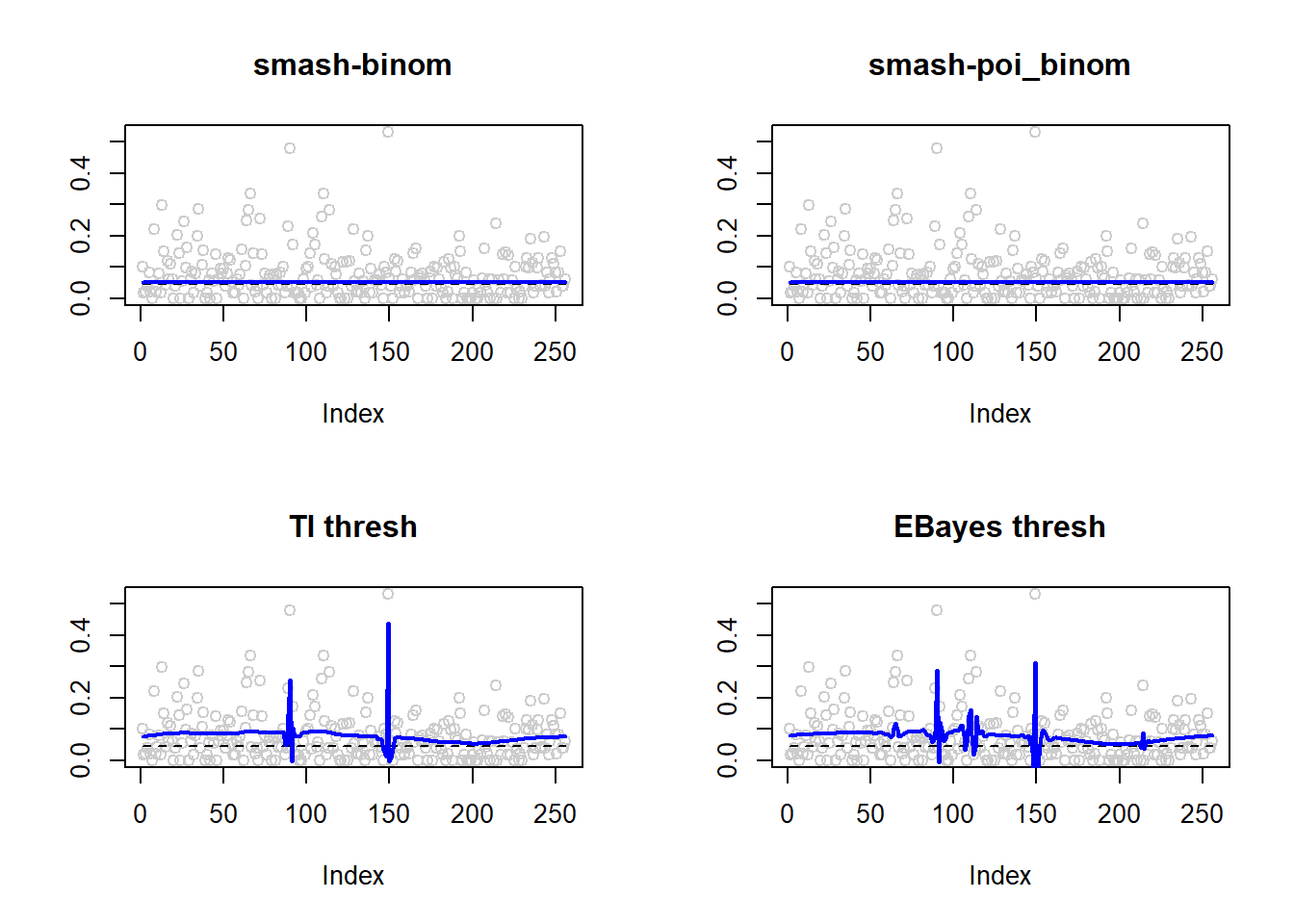
Expand here to see past versions of unnamed-chunk-4-3.png:
| Version | Author | Date |
|---|---|---|
| 9d6ca07 | Dongyue | 2018-05-18 |
As expected, when \(n\) is large and \(p\) is small, Poisson distribution is a good approximation to binomial distribution.
\(ntri\) small, \(p\) small
n=256
p=rep(-3,n)
set.seed(111)
ntri=rpois(n,5)+1
result=simu_study(p,ntri=ntri)
ggplot(df2gg(result$err),aes(x=method,y=MSE))+geom_boxplot(aes(fill=method))+labs(x='')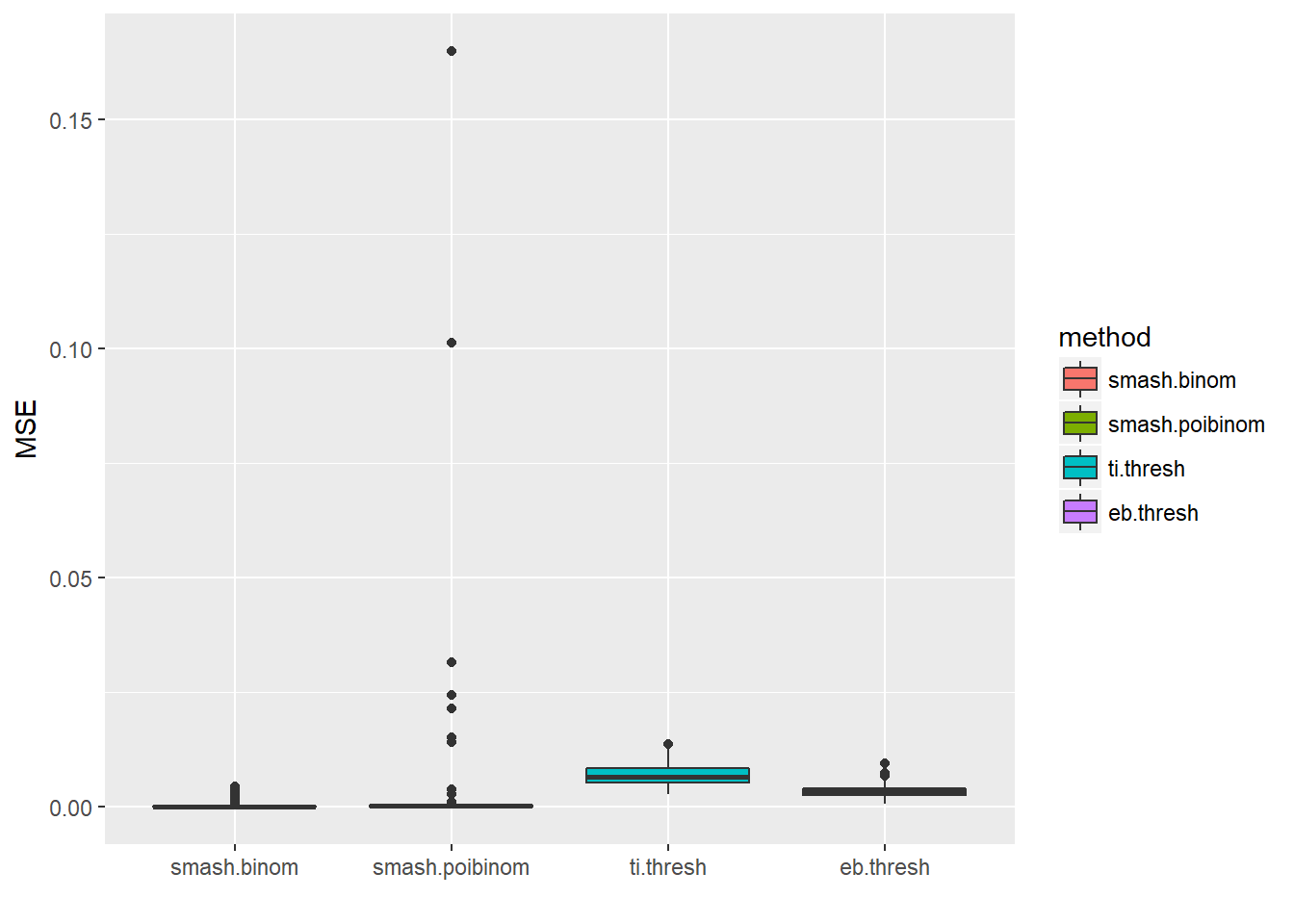
Expand here to see past versions of unnamed-chunk-5-1.png:
| Version | Author | Date |
|---|---|---|
| 9d6ca07 | Dongyue | 2018-05-18 |
apply(result$err,2,mean) smash.binom smash.poibinom ti.thresh eb.thresh
0.0004194395 0.0039619590 0.0068907435 0.0034840933 par(mfrow=c(2,2))
plot(result$est$x/ntri,col='gray80',ylab='',main='smash-binom')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$smash.binom.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='smash-poi_binom')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$smash.poibinom.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='TI thresh')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$ti.thresh.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='EBayes thresh')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$eb.thresh.out,col=4,lwd=2)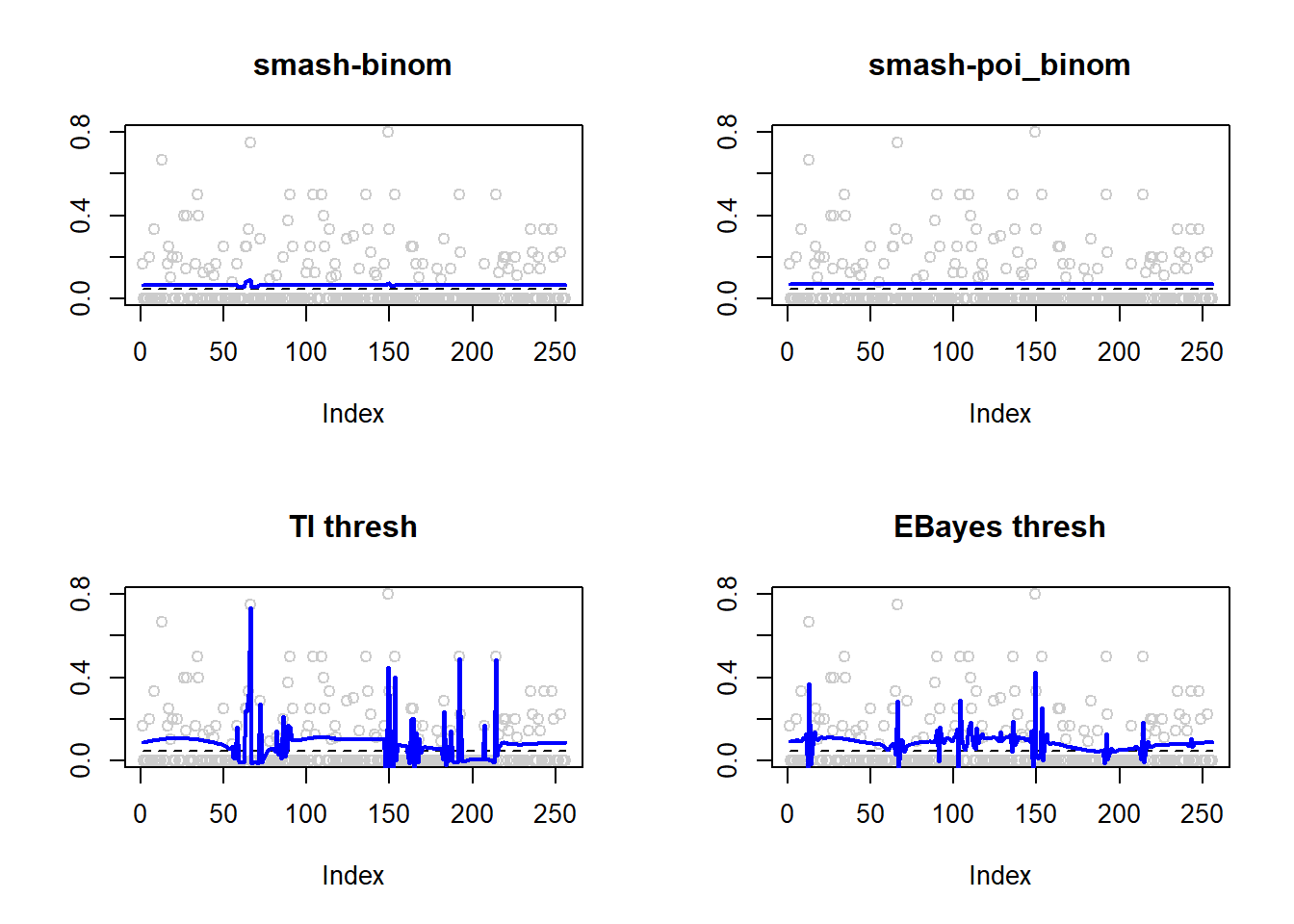
Expand here to see past versions of unnamed-chunk-5-2.png:
| Version | Author | Date |
|---|---|---|
| 9d6ca07 | Dongyue | 2018-05-18 |
Steps
Small \(n\)
p=c(rep(-2,64), rep(0, 64), rep(2, 64), rep(-2, 64))
set.seed(111)
ntri=rpois(256,5)+1
result=simu_study(p,ntri=ntri)
ggplot(df2gg(result$err),aes(x=method,y=MSE))+geom_boxplot(aes(fill=method))+labs(x='')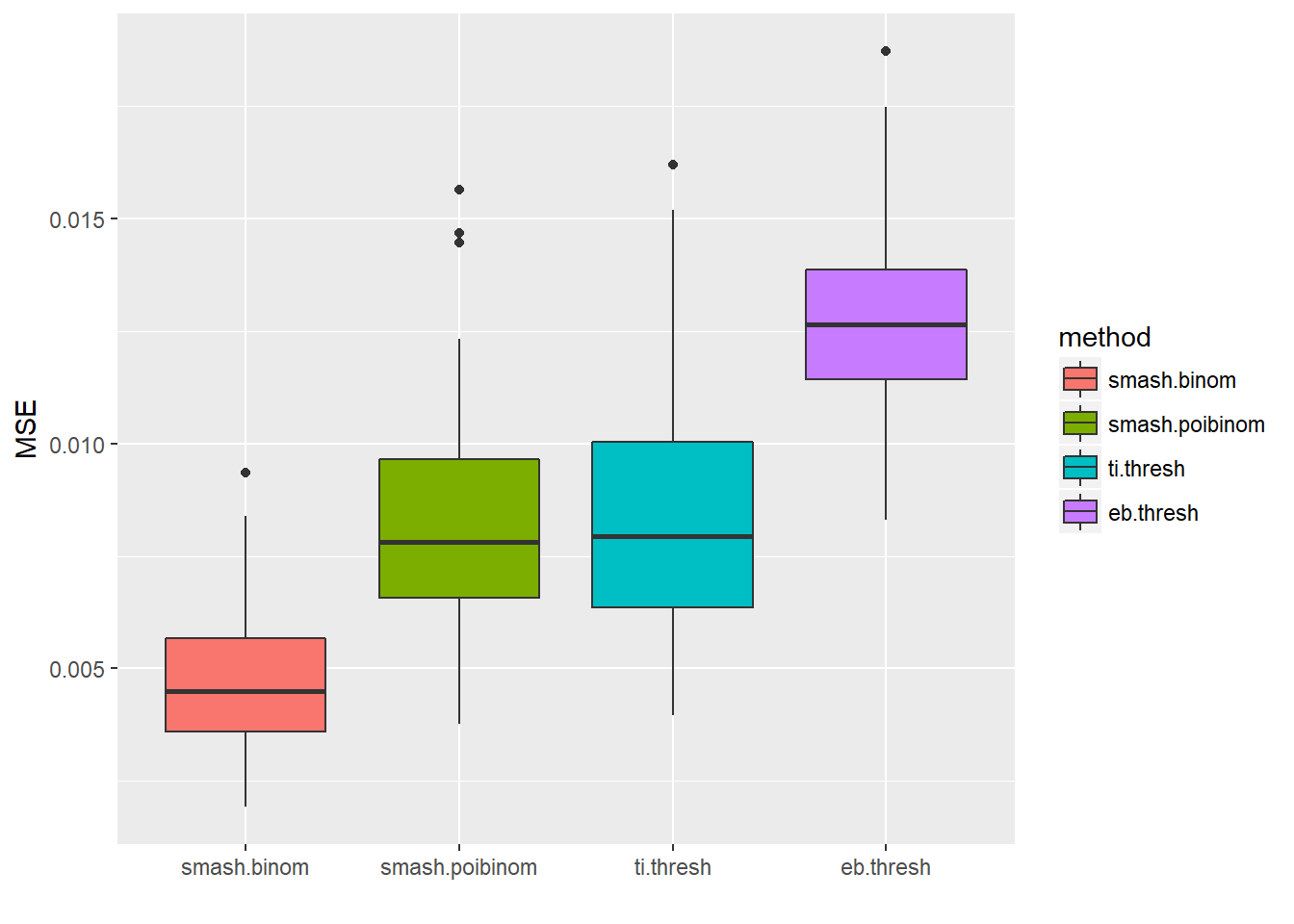
Expand here to see past versions of unnamed-chunk-6-1.png:
| Version | Author | Date |
|---|---|---|
| 9d6ca07 | Dongyue | 2018-05-18 |
apply(result$err,2,mean) smash.binom smash.poibinom ti.thresh eb.thresh
0.004713551 0.008199597 0.008406726 0.012840545 par(mfrow=c(2,2))
plot(result$est$x/ntri,col='gray80',ylab='',main='smash-binom')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$smash.binom.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='smash-poi_binom')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$smash.poibinom.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='TI thresh')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$ti.thresh.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='EBayes thresh')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$eb.thresh.out,col=4,lwd=2)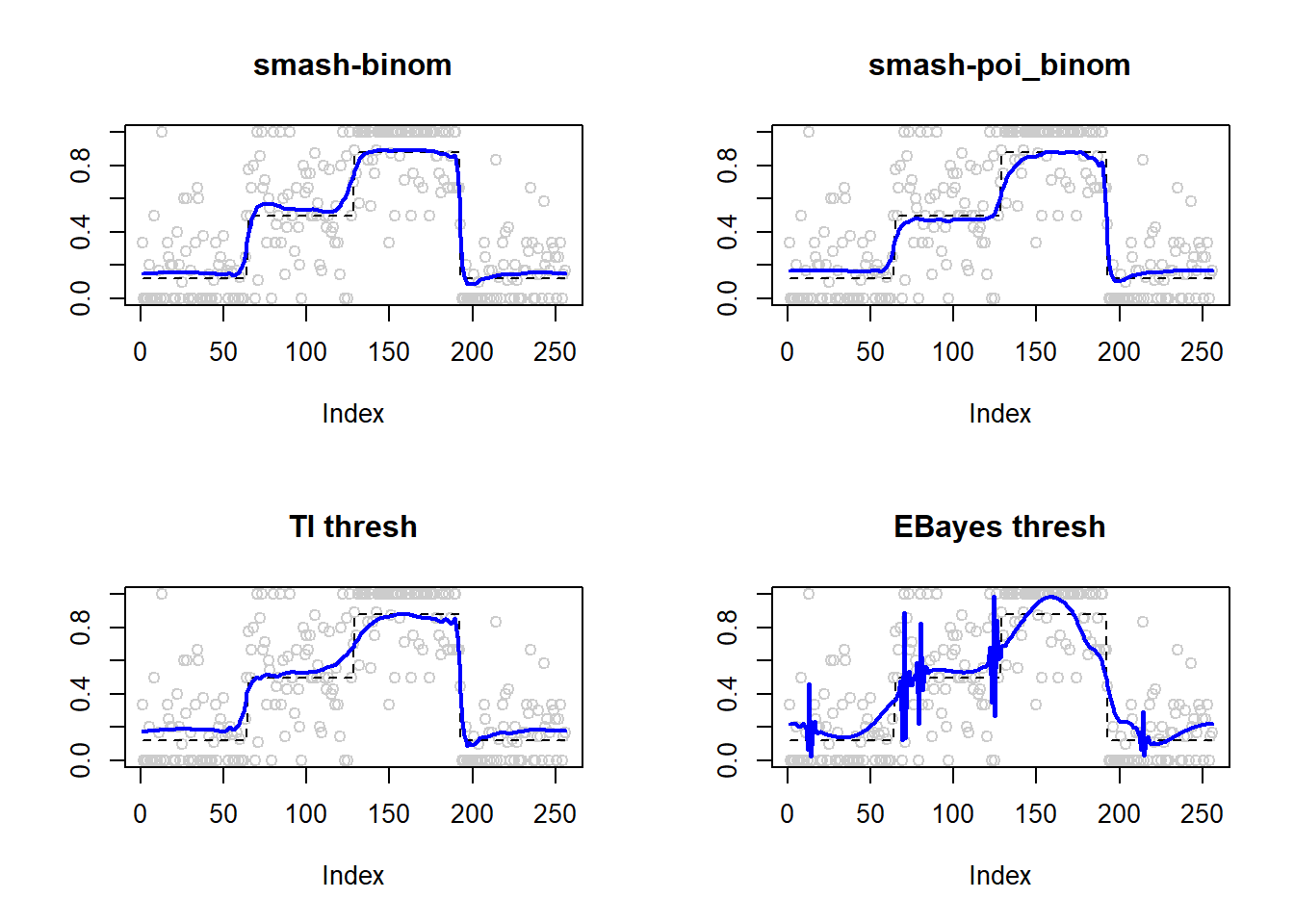
Expand here to see past versions of unnamed-chunk-6-2.png:
| Version | Author | Date |
|---|---|---|
| 9d6ca07 | Dongyue | 2018-05-18 |
Large \(n\)
p=c(rep(-2,64), rep(0, 64), rep(2, 64), rep(-2, 64))
set.seed(111)
ntri=ntri+44
result=simu_study(p,ntri=ntri)
ggplot(df2gg(result$err),aes(x=method,y=MSE))+geom_boxplot(aes(fill=method))+labs(x='')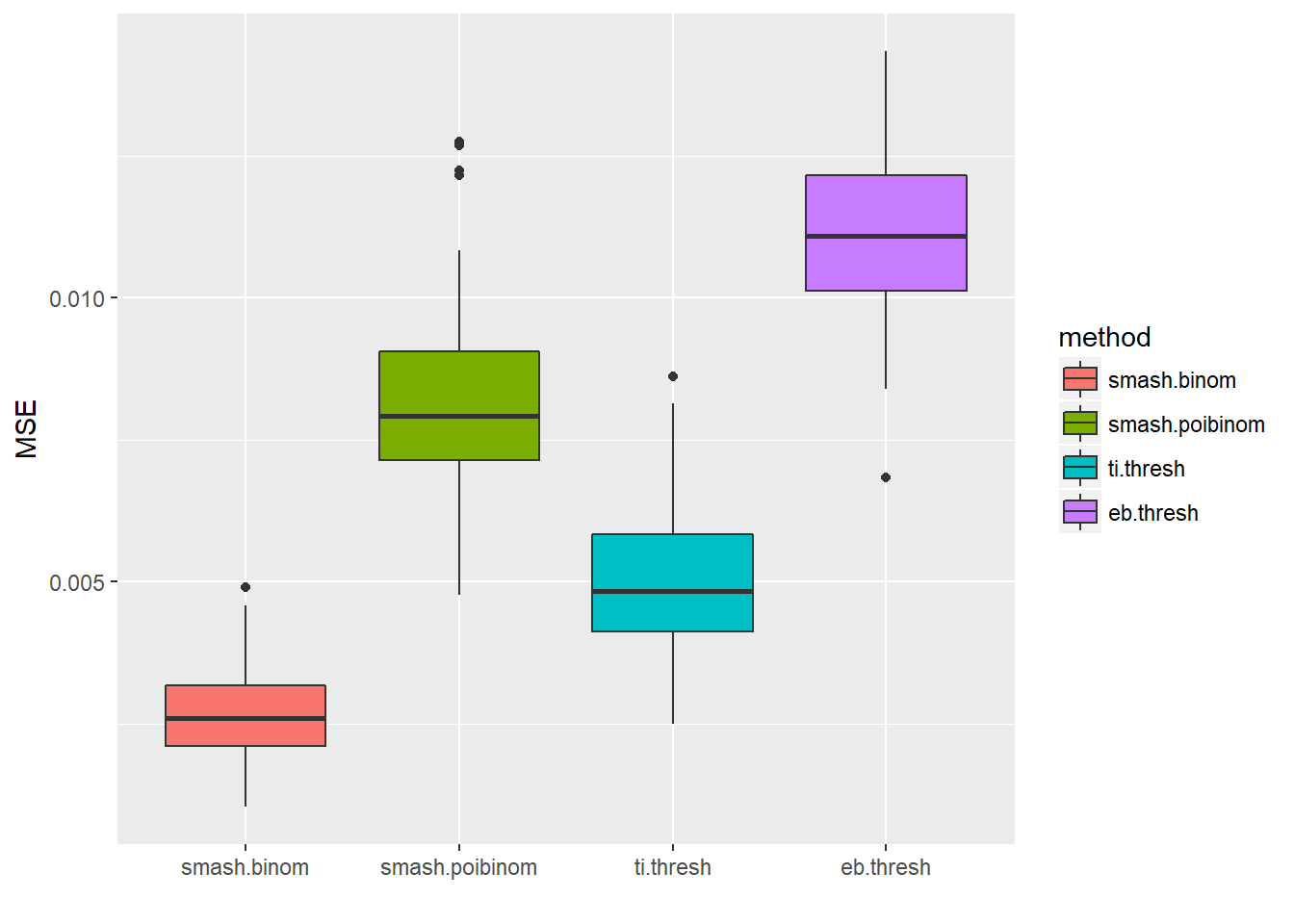
Expand here to see past versions of unnamed-chunk-7-1.png:
| Version | Author | Date |
|---|---|---|
| 9d6ca07 | Dongyue | 2018-05-18 |
apply(result$err,2,mean) smash.binom smash.poibinom ti.thresh eb.thresh
0.002653184 0.008115945 0.005058030 0.011180475 par(mfrow=c(2,2))
plot(result$est$x/ntri,col='gray80',ylab='',main='smash-binom')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$smash.binom.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='smash-poi_binom')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$smash.poibinom.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='TI thresh')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$ti.thresh.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='EBayes thresh')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$eb.thresh.out,col=4,lwd=2)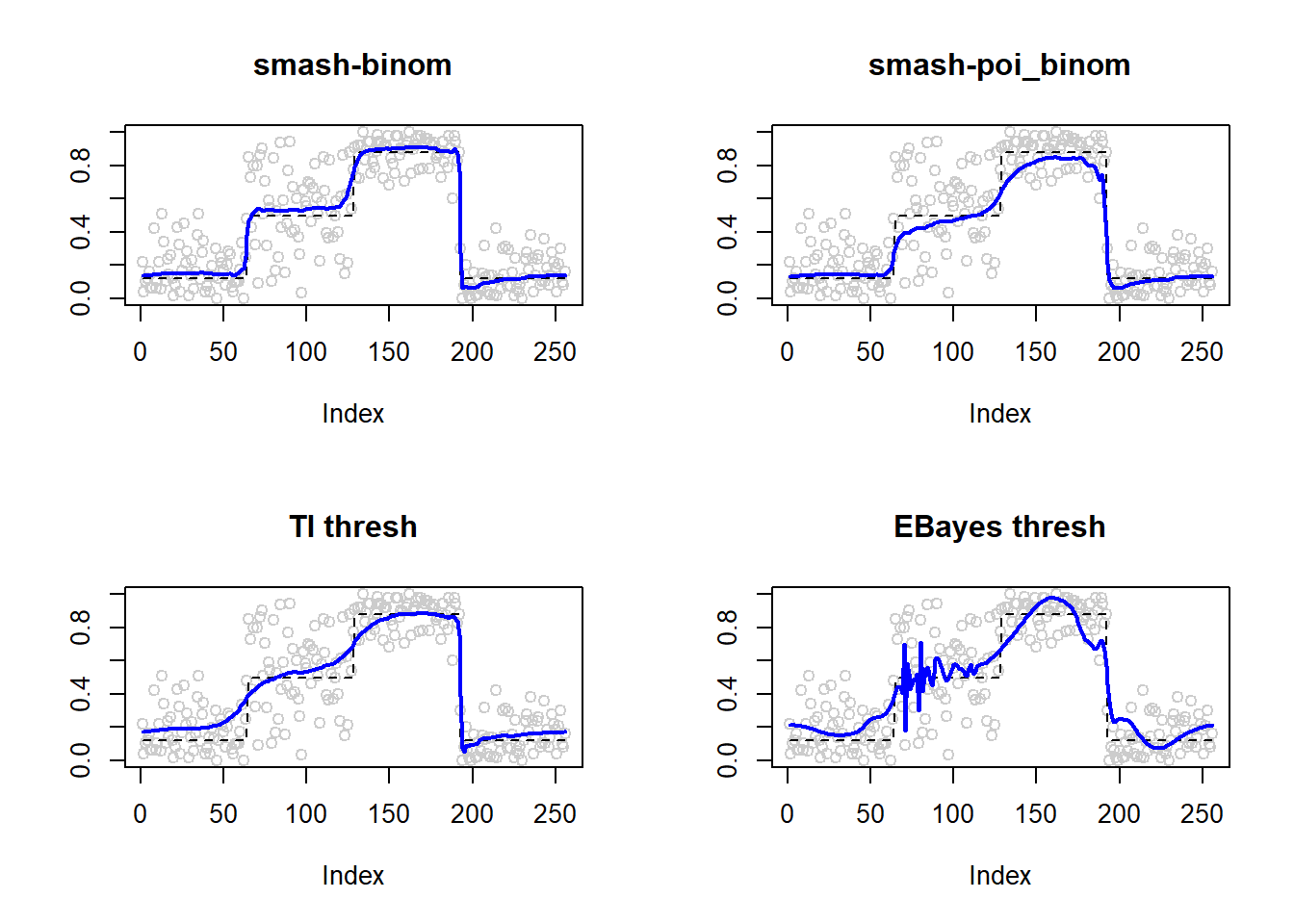
Expand here to see past versions of unnamed-chunk-7-2.png:
| Version | Author | Date |
|---|---|---|
| 9d6ca07 | Dongyue | 2018-05-18 |
Bumps
Small \(n\)
set.seed(111)
ntri=rpois(256,5)+1
result=simu_study(p,ntri=ntri)
ggplot(df2gg(result$err),aes(x=method,y=MSE))+geom_boxplot(aes(fill=method))+labs(x='')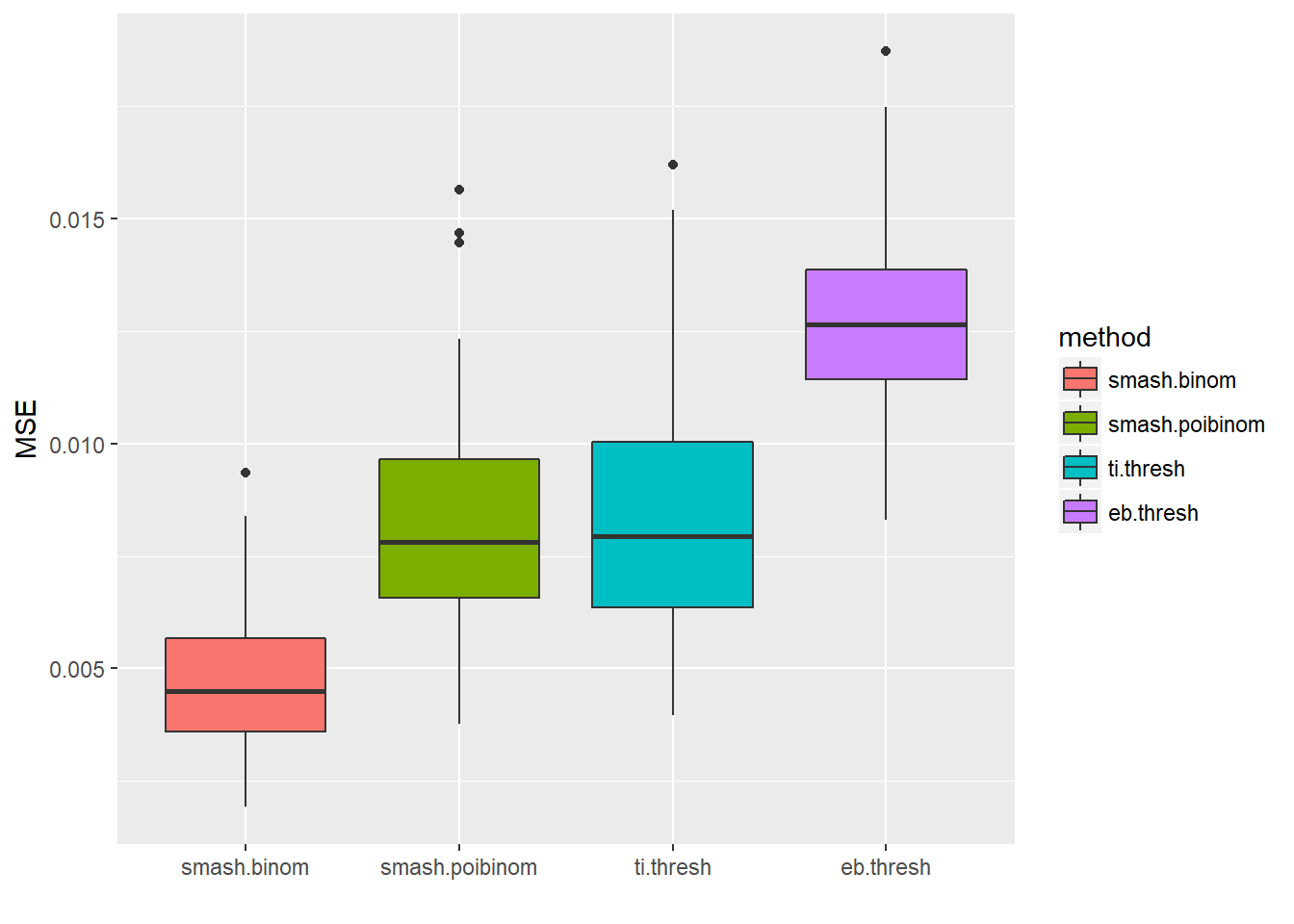
Expand here to see past versions of unnamed-chunk-8-1.png:
| Version | Author | Date |
|---|---|---|
| 9d6ca07 | Dongyue | 2018-05-18 |
apply(result$err,2,mean) smash.binom smash.poibinom ti.thresh eb.thresh
0.004713551 0.008199597 0.008406726 0.012840545 par(mfrow=c(2,2))
plot(result$est$x/ntri,col='gray80',ylab='',main='smash-binom')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$smash.binom.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='smash-poi_binom')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$smash.poibinom.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='TI thresh')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$ti.thresh.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='EBayes thresh')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$eb.thresh.out,col=4,lwd=2)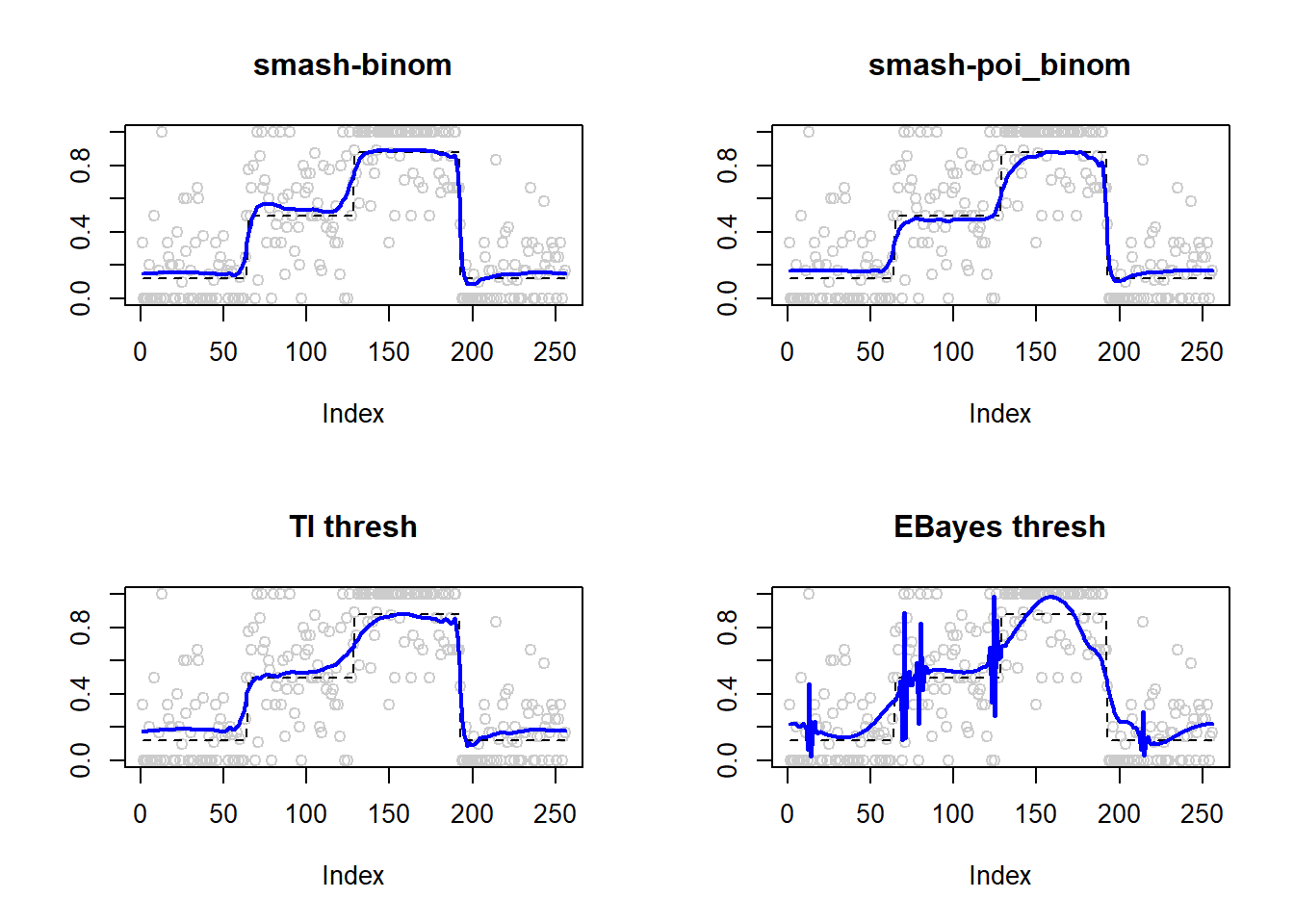
Expand here to see past versions of unnamed-chunk-8-2.png:
| Version | Author | Date |
|---|---|---|
| 9d6ca07 | Dongyue | 2018-05-18 |
Large \(n\).
m=seq(0,1,length.out = 256)
h = c(4, 5, 3, 4, 5, 4.2, 2.1, 4.3, 3.1, 5.1, 4.2)
w = c(0.005, 0.005, 0.006, 0.01, 0.01, 0.03, 0.01, 0.01, 0.005,0.008,0.005)
t=c(.1,.13,.15,.23,.25,.4,.44,.65,.76,.78,.81)
f = c()
for(i in 1:length(m)){
f[i]=sum(h*(1+((m[i]-t)/w)^4)^(-1))
}
p=f-3
set.seed(111)
ntri=ntri+44
result=simu_study(p,ntri=ntri)
ggplot(df2gg(result$err),aes(x=method,y=MSE))+geom_boxplot(aes(fill=method))+labs(x='')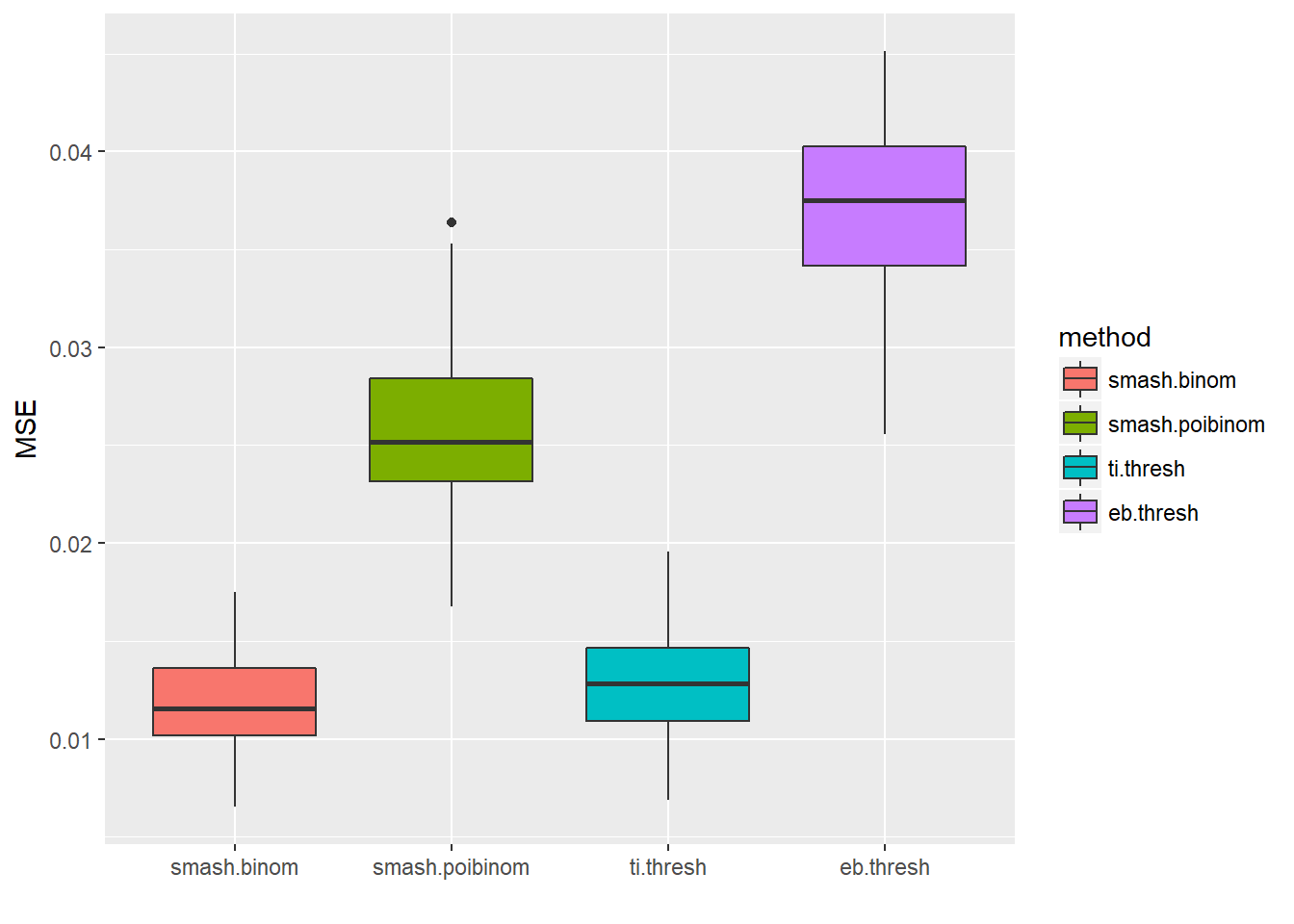
Expand here to see past versions of unnamed-chunk-9-1.png:
| Version | Author | Date |
|---|---|---|
| 9d6ca07 | Dongyue | 2018-05-18 |
apply(result$err,2,mean) smash.binom smash.poibinom ti.thresh eb.thresh
0.01186805 0.02590525 0.01273087 0.03682167 par(mfrow=c(2,2))
plot(result$est$x/ntri,col='gray80',ylab='',main='smash-binom')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$smash.binom.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='smash-poi_binom')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$smash.poibinom.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='TI thresh')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$ti.thresh.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='EBayes thresh')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$eb.thresh.out,col=4,lwd=2)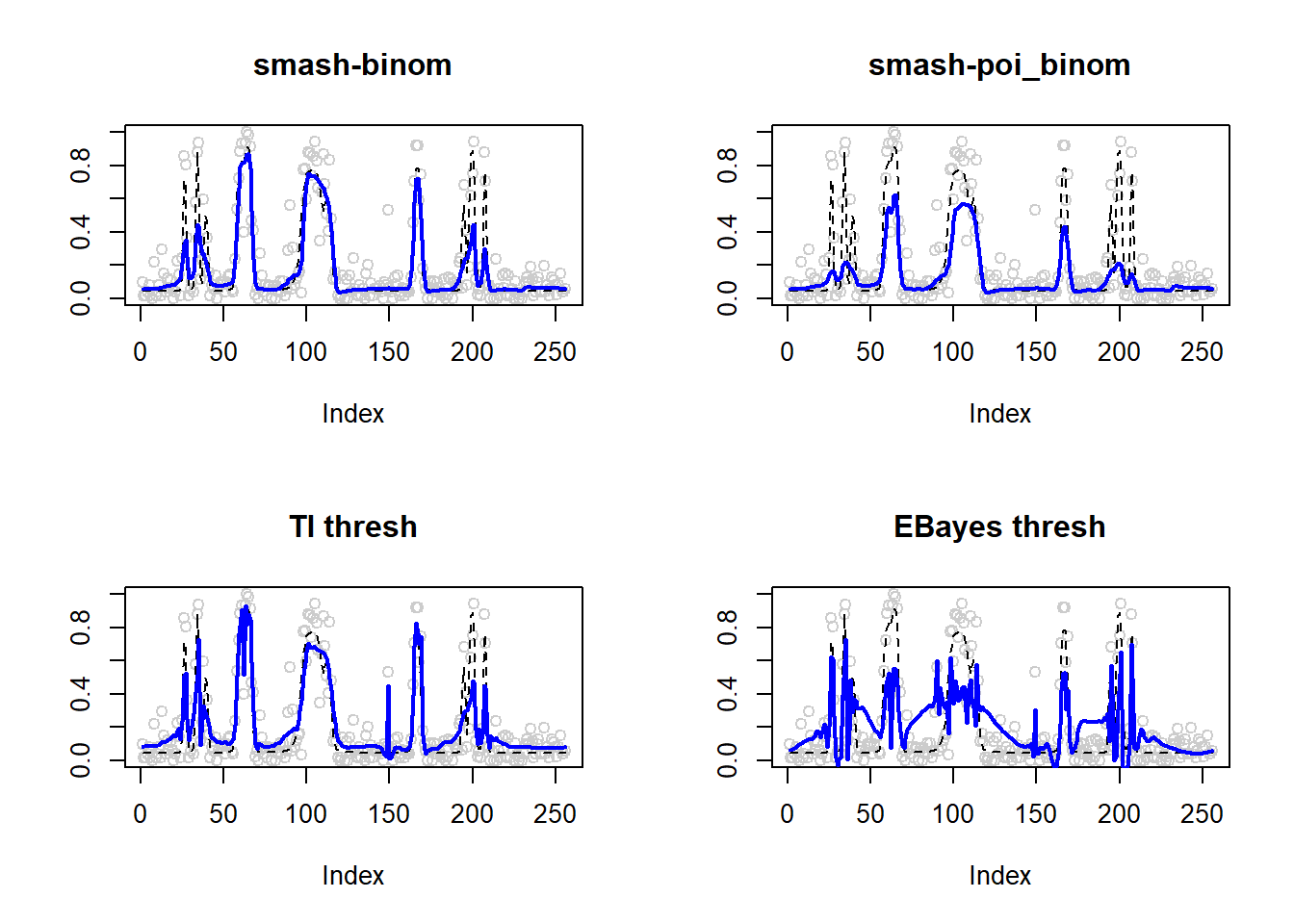
Expand here to see past versions of unnamed-chunk-9-2.png:
| Version | Author | Date |
|---|---|---|
| 9d6ca07 | Dongyue | 2018-05-18 |
Spike mean
Small \(n\)
set.seed(111)
ntri=rpois(256,5)+1
result=simu_study(p,ntri=ntri)
ggplot(df2gg(result$err),aes(x=method,y=MSE))+geom_boxplot(aes(fill=method))+labs(x='')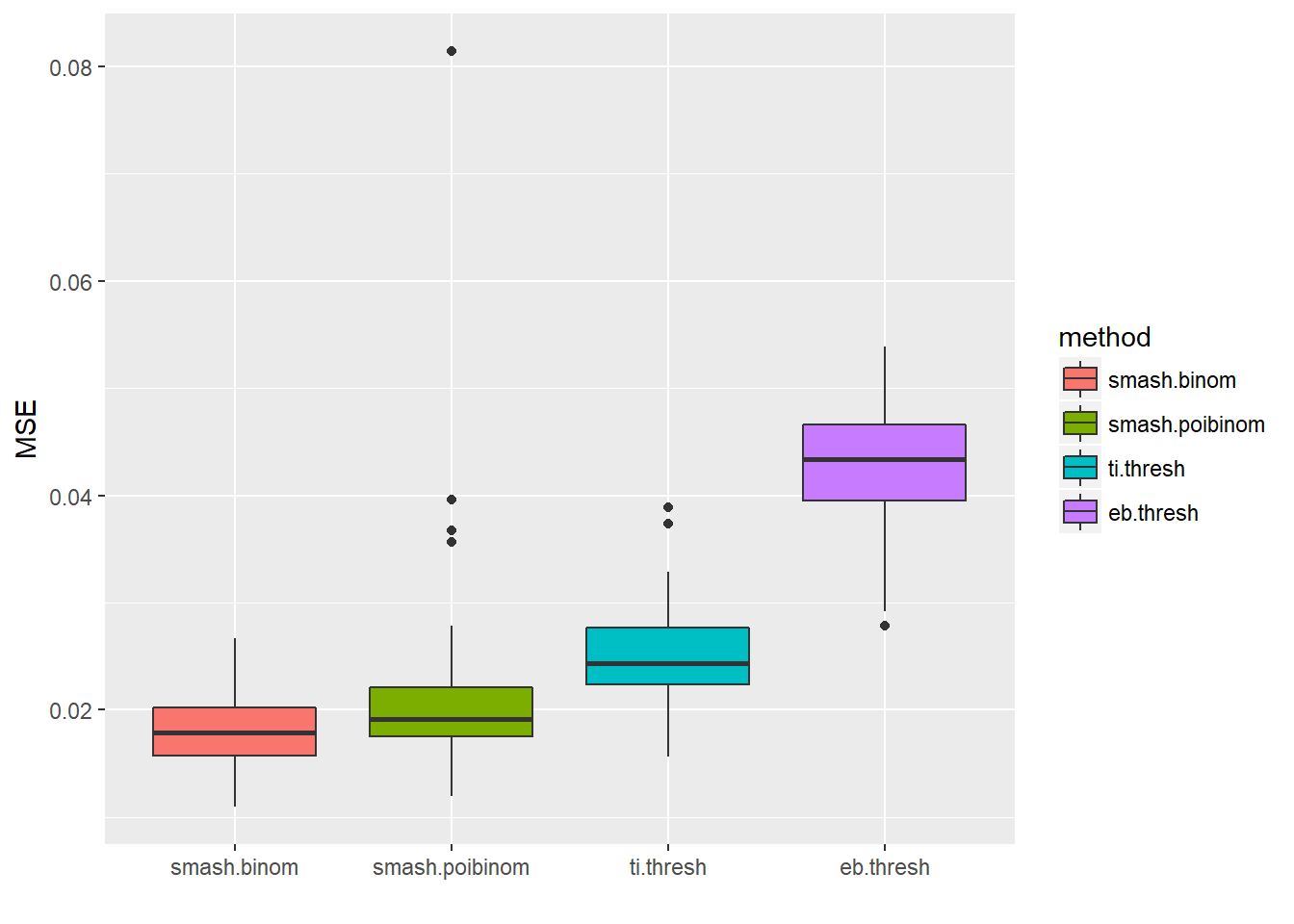
Expand here to see past versions of unnamed-chunk-10-1.png:
| Version | Author | Date |
|---|---|---|
| 9d6ca07 | Dongyue | 2018-05-18 |
apply(result$err,2,mean) smash.binom smash.poibinom ti.thresh eb.thresh
0.01793979 0.02080153 0.02484011 0.04274885 par(mfrow=c(2,2))
plot(result$est$x/ntri,col='gray80',ylab='',main='smash-binom')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$smash.binom.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='smash-poi_binom')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$smash.poibinom.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='TI thresh')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$ti.thresh.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='EBayes thresh')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$eb.thresh.out,col=4,lwd=2)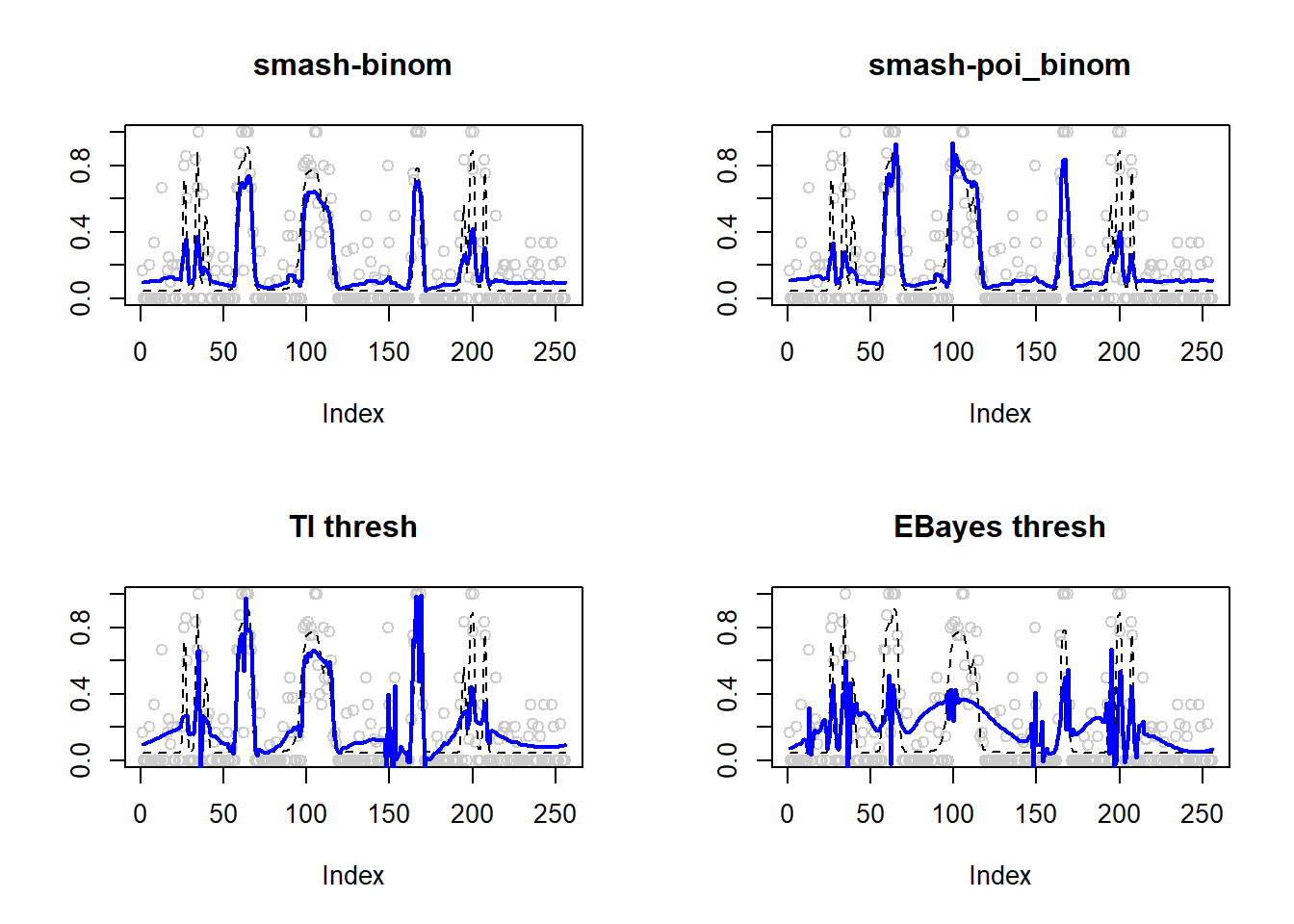
Expand here to see past versions of unnamed-chunk-10-2.png:
| Version | Author | Date |
|---|---|---|
| 9d6ca07 | Dongyue | 2018-05-18 |
Large \(n\)
spike.f = function(x) (0.75 * exp(-500 * (x - 0.23)^2) + 1.5 * exp(-2000 * (x - 0.33)^2) + 3 * exp(-8000 * (x - 0.47)^2) + 2.25 * exp(-16000 *
(x - 0.69)^2) + 0.5 * exp(-32000 * (x - 0.83)^2))
n = 256
t = 1:n/n
p = spike.f(t)*2-2
set.seed(111)
ntri=ntri+44
result=simu_study(p,ntri=ntri)
ggplot(df2gg(result$err),aes(x=method,y=MSE))+geom_boxplot(aes(fill=method))+labs(x='')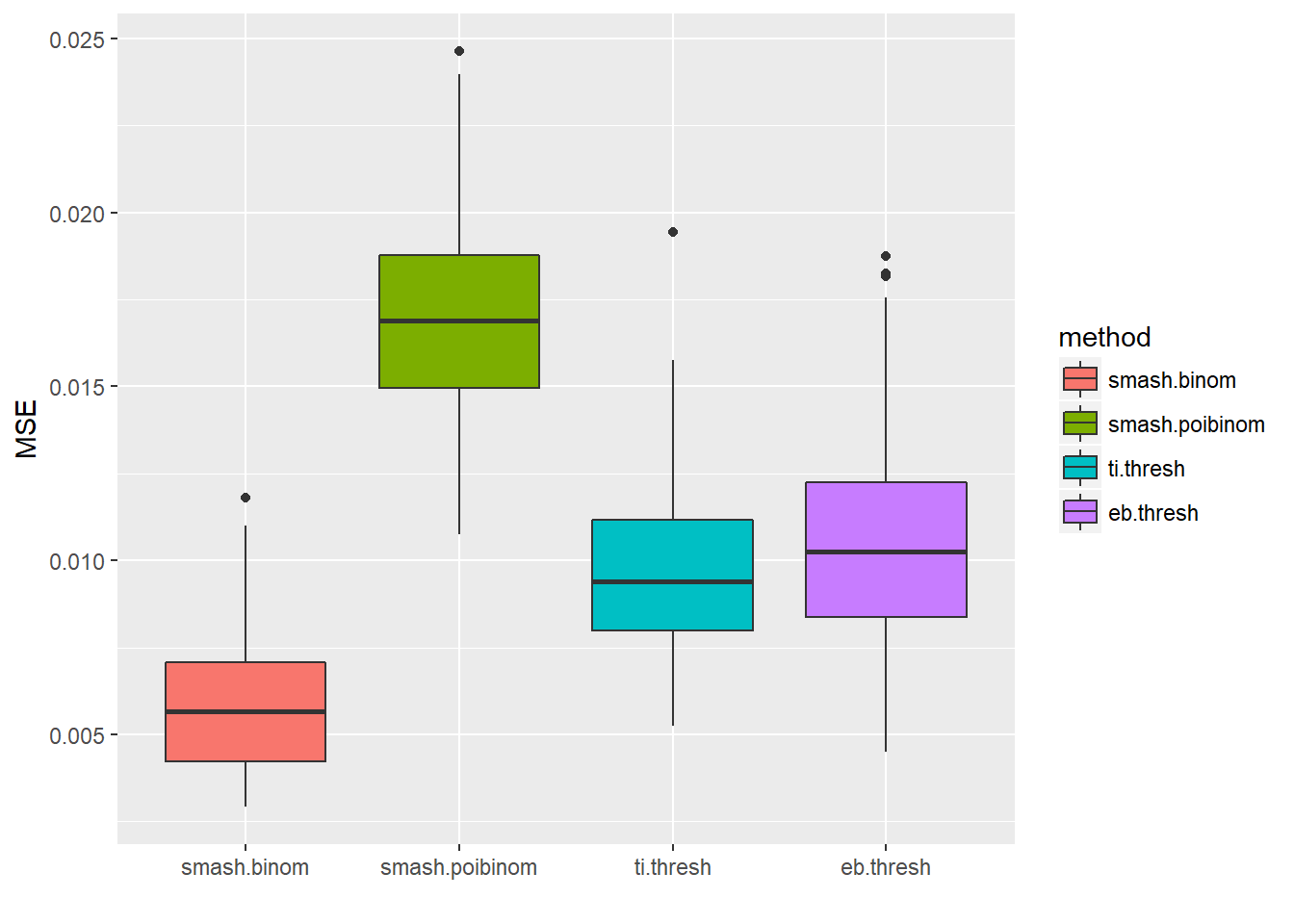
Expand here to see past versions of unnamed-chunk-11-1.png:
| Version | Author | Date |
|---|---|---|
| 9d6ca07 | Dongyue | 2018-05-18 |
apply(result$err,2,mean) smash.binom smash.poibinom ti.thresh eb.thresh
0.005917600 0.017084435 0.009726599 0.010659236 par(mfrow=c(2,2))
plot(result$est$x/ntri,col='gray80',ylab='',main='smash-binom')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$smash.binom.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='smash-poi_binom')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$smash.poibinom.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='TI thresh')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$ti.thresh.out,col=4,lwd=2)
plot(result$est$x/ntri,col='gray80',ylab='',main='EBayes thresh')
lines(exp(p)/(1+exp(p)),lty=2)
lines(result$est$eb.thresh.out,col=4,lwd=2)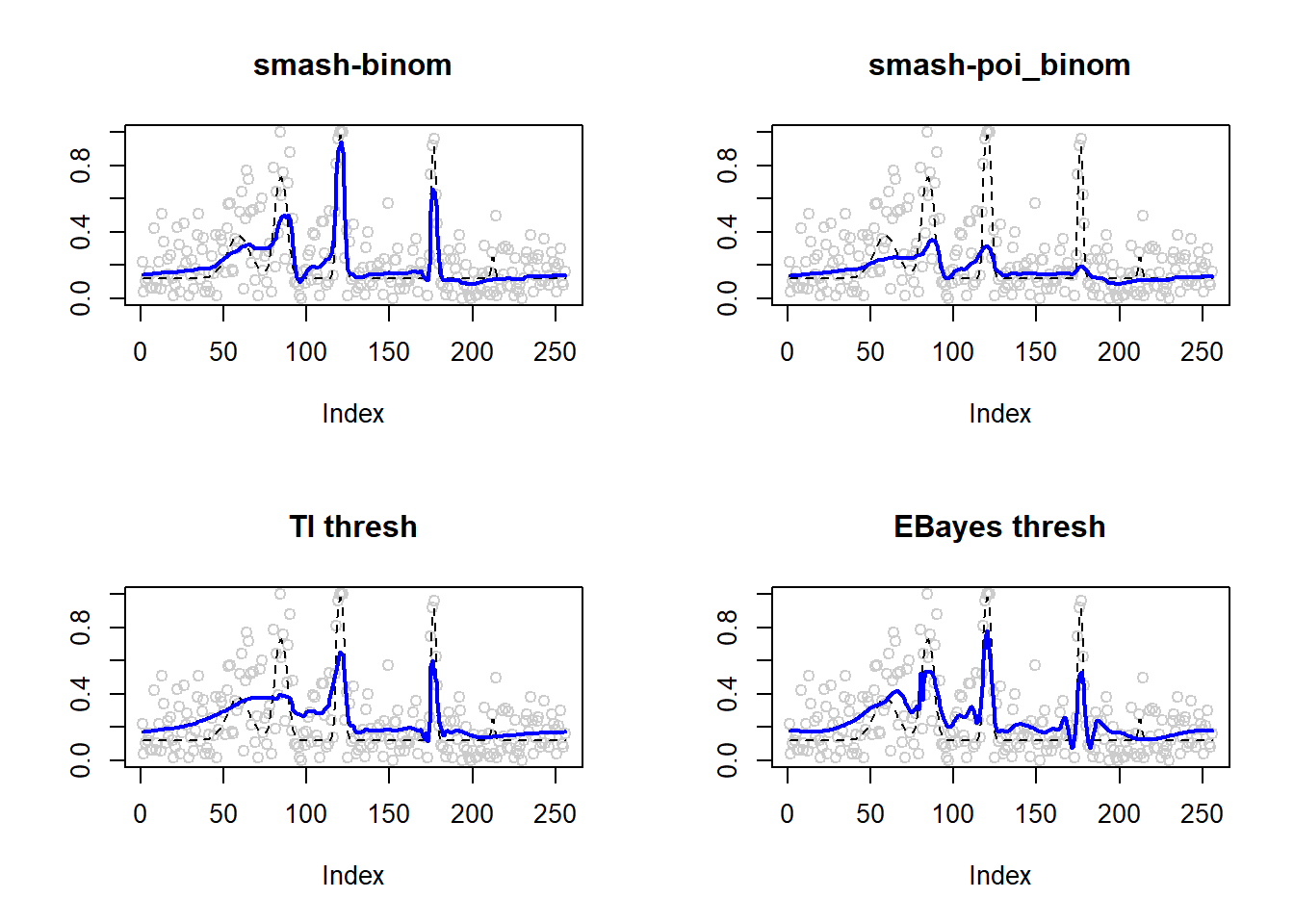
Expand here to see past versions of unnamed-chunk-11-2.png:
| Version | Author | Date |
|---|---|---|
| 9d6ca07 | Dongyue | 2018-05-18 |
Summary
When \(p\) is unchanged, smaller number of trials lead to larger MSE for smashgen-binom, TI thresh and EB thresh, while this is not the case for smash-poi_binom. This seems strange. I think the reason is that with the increase of \(ntri\), the variance also increases. Hence, when we treat the data as poisson and try to use ash ‘estimating’ the \(\lambda_i\)(or \(n_ip_i\)), the estimates are not as good as the smaller variance cases.
Session information
sessionInfo()R version 3.4.0 (2017-04-21)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 16299)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.1252
[2] LC_CTYPE=English_United States.1252
[3] LC_MONETARY=English_United States.1252
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] EbayesThresh_1.4-12 ggplot2_2.2.1 smashrgen_0.1.0
[4] wavethresh_4.6.8 MASS_7.3-47 caTools_1.17.1
[7] ashr_2.2-7 smashr_1.1-5
loaded via a namespace (and not attached):
[1] Rcpp_0.12.16 plyr_1.8.4 compiler_3.4.0
[4] git2r_0.21.0 workflowr_1.0.1 R.methodsS3_1.7.1
[7] R.utils_2.6.0 bitops_1.0-6 iterators_1.0.8
[10] tools_3.4.0 digest_0.6.13 tibble_1.3.3
[13] evaluate_0.10 gtable_0.2.0 lattice_0.20-35
[16] rlang_0.1.2 Matrix_1.2-9 foreach_1.4.3
[19] yaml_2.1.19 parallel_3.4.0 stringr_1.3.0
[22] knitr_1.20 REBayes_1.3 rprojroot_1.3-2
[25] grid_3.4.0 data.table_1.10.4-3 rmarkdown_1.8
[28] magrittr_1.5 whisker_0.3-2 backports_1.0.5
[31] scales_0.4.1 codetools_0.2-15 htmltools_0.3.5
[34] assertthat_0.2.0 colorspace_1.3-2 labeling_0.3
[37] stringi_1.1.6 Rmosek_8.0.69 lazyeval_0.2.1
[40] munsell_0.4.3 doParallel_1.0.11 pscl_1.4.9
[43] truncnorm_1.0-7 SQUAREM_2017.10-1 R.oo_1.21.0 This reproducible R Markdown analysis was created with workflowr 1.0.1