Differential transcript detection
Philipp Ross
2018-09-25
Last updated: 2018-10-07
Code version: 33d7a01
What transcripts can we detect in one strain, but not the others?
First we need to detect transcript differences. To do this we run the detect_transcripts.R script found within code/differential_detection directory.
Are these genes enriched for any GO terms?
Next we run the topGO script:
Are undetected genes sometimes due to known polymorphic regions?
We can address this by calculating the coverage across each exon and comparing the fraction covered by reads.
for i in $(seq 7);
do bedtools coverage -a -b ../data/bam/3d7.3d7_v3_chr.tp${i}.bam -s -split > ../output/differential_detection/coverages/3d7_tp${i}_cov.tsv;
done
for strain in hb3 it;
do for i in $(seq 7);
do bedtools coverage -a -b ../data/bam/${strain}.3d7chr.tp${i}.bam -s -split > ../output/differential_detection/coverages/${strain}_tp${i}_cov.tsv;
done
doneread_cov <- function(file) {
df <- readr::read_tsv(file,col_names=F) %>%
dplyr::select(X9,X10,X11,X12,X13) %>%
dplyr::rename(att=X9,reads=X10,nonzero=X11,len=X12,fraction=X13)
df$exon <- apply(df, 1, function(x) {
stringr::str_replace(stringr::str_split(stringr::str_split(x[["att"]],";")[[1]][1],"=")[[1]][2],"exon_","")
})
return(dplyr::select(df,-att))
}
files_3d7 <- list.files("../output/differential_detection/coverages/",pattern="3d7",full.names=T)
files_hb3 <- list.files("../output/differential_detection/coverages/",pattern="hb3",full.names=T)
files_it <- list.files("../output/differential_detection/coverages/",pattern="it",full.names=T)
tp1_3d7 <- read_cov(files_3d7[1]) %>% mutate(strain="3D7",tp="T1")
tp2_3d7 <- read_cov(files_3d7[2]) %>% mutate(strain="3D7",tp="T2")
tp3_3d7 <- read_cov(files_3d7[3]) %>% mutate(strain="3D7",tp="T3")
tp4_3d7 <- read_cov(files_3d7[4]) %>% mutate(strain="3D7",tp="T4")
tp5_3d7 <- read_cov(files_3d7[5]) %>% mutate(strain="3D7",tp="T5")
tp6_3d7 <- read_cov(files_3d7[6]) %>% mutate(strain="3D7",tp="T6")
tp7_3d7 <- read_cov(files_3d7[7]) %>% mutate(strain="3D7",tp="T7")
df_3d7 <- rbind(tp1_3d7,tp2_3d7,tp3_3d7,tp4_3d7,tp5_3d7,tp6_3d7,tp7_3d7)
rm(tp1_3d7,tp2_3d7,tp3_3d7,tp4_3d7,tp5_3d7,tp6_3d7,tp7_3d7)
tp1_hb3 <- read_cov(files_hb3[1]) %>% mutate(strain="HB3",tp="T1")
tp2_hb3 <- read_cov(files_hb3[2]) %>% mutate(strain="HB3",tp="T2")
tp3_hb3 <- read_cov(files_hb3[3]) %>% mutate(strain="HB3",tp="T3")
tp4_hb3 <- read_cov(files_hb3[4]) %>% mutate(strain="HB3",tp="T4")
tp5_hb3 <- read_cov(files_hb3[5]) %>% mutate(strain="HB3",tp="T5")
tp6_hb3 <- read_cov(files_hb3[6]) %>% mutate(strain="HB3",tp="T6")
tp7_hb3 <- read_cov(files_hb3[7]) %>% mutate(strain="HB3",tp="T7")
df_hb3 <- rbind(tp1_hb3,tp2_hb3,tp3_hb3,tp4_hb3,tp5_hb3,tp6_hb3,tp7_hb3)
rm(tp1_hb3,tp2_hb3,tp3_hb3,tp4_hb3,tp5_hb3,tp6_hb3,tp7_hb3)
tp1_it <- read_cov(files_it[1]) %>% mutate(strain="IT",tp="T1")
tp2_it <- read_cov(files_it[2]) %>% mutate(strain="IT",tp="T2")
tp3_it <- read_cov(files_it[3]) %>% mutate(strain="IT",tp="T3")
tp4_it <- read_cov(files_it[4]) %>% mutate(strain="IT",tp="T4")
tp5_it <- read_cov(files_it[5]) %>% mutate(strain="IT",tp="T5")
tp6_it <- read_cov(files_it[6]) %>% mutate(strain="IT",tp="T6")
tp7_it <- read_cov(files_it[7]) %>% mutate(strain="IT",tp="T7")
df_it <- rbind(tp1_it,tp2_it,tp3_it,tp4_it,tp5_it,tp6_it,tp7_it)
rm(tp1_it,tp2_it,tp3_it,tp4_it,tp5_it,tp6_it,tp7_it)
coverages <- rbind(df_3d7,df_hb3,df_it)
coverages <- coverages[,c(5,6,7,1,2,3,4)] %>%
separate(exon, into = c("gene_id","exon"), sep = "-") %>%
group_by(gene_id,strain,tp) %>%
summarise(reads=sum(reads),nonzero=sum(nonzero),len=sum(len),fraction=sum(nonzero)/sum(len))
readr::write_tsv(x=coverages,path="../output/differential_detection/coverages/coverages.tsv")coverages$tp <- factor(coverages$tp, levels = c("T1","T2","T3","T4","T5","T6","T7"))
core_genes <- read_tsv("../data/gene_lists/core_pf3d7_genes.txt",col_names=F)$X1
mean_coverages <- coverages %>%
group_by(gene_id,strain) %>%
summarise(fraction=mean(fraction)) %>%
spread(strain,fraction) %>%
mutate(diffhb3 = `3D7` - HB3, diffit = `3D7` - IT)
max_coverages <- coverages %>%
group_by(gene_id,strain) %>%
summarise(fraction=max(fraction)) %>%
spread(strain,fraction) %>%
mutate(diffhb3 = `3D7` - HB3, diffit = `3D7` - IT)
g1 <- ggplot(mean_coverages,aes(x=diffhb3)) +
geom_histogram(color="grey90") +
xlab("Difference from HB3")
g2 <- ggplot(mean_coverages,aes(x=diffit)) +
geom_histogram(color="grey90") +
xlab("Difference from IT")
p <- plot_grid(g1,g2,nrow=1)
print(p)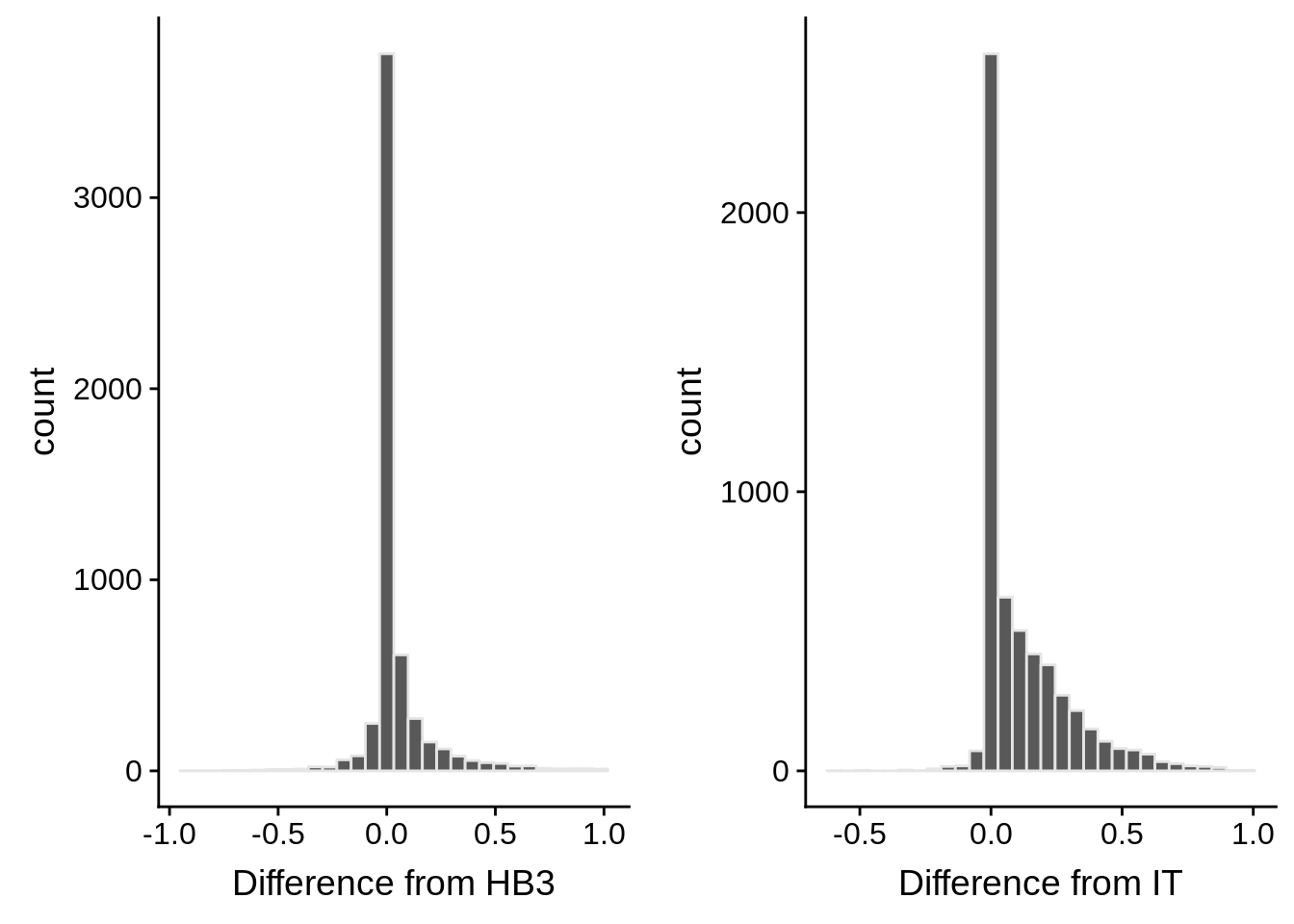
g <- ggplot(mean_coverages,aes(x=`3D7`,y=HB3)) + geom_point() + geom_point(data=mean_coverages %>% dplyr::filter(gene_id == "PF3D7_1222600"),color = "#E41A1C") + geom_text(data=mean_coverages %>% dplyr::filter(gene_id == "PF3D7_1222600"),label="PF3D7_1222600", color = "#E41A1C", hjust = 1.1)
print(g)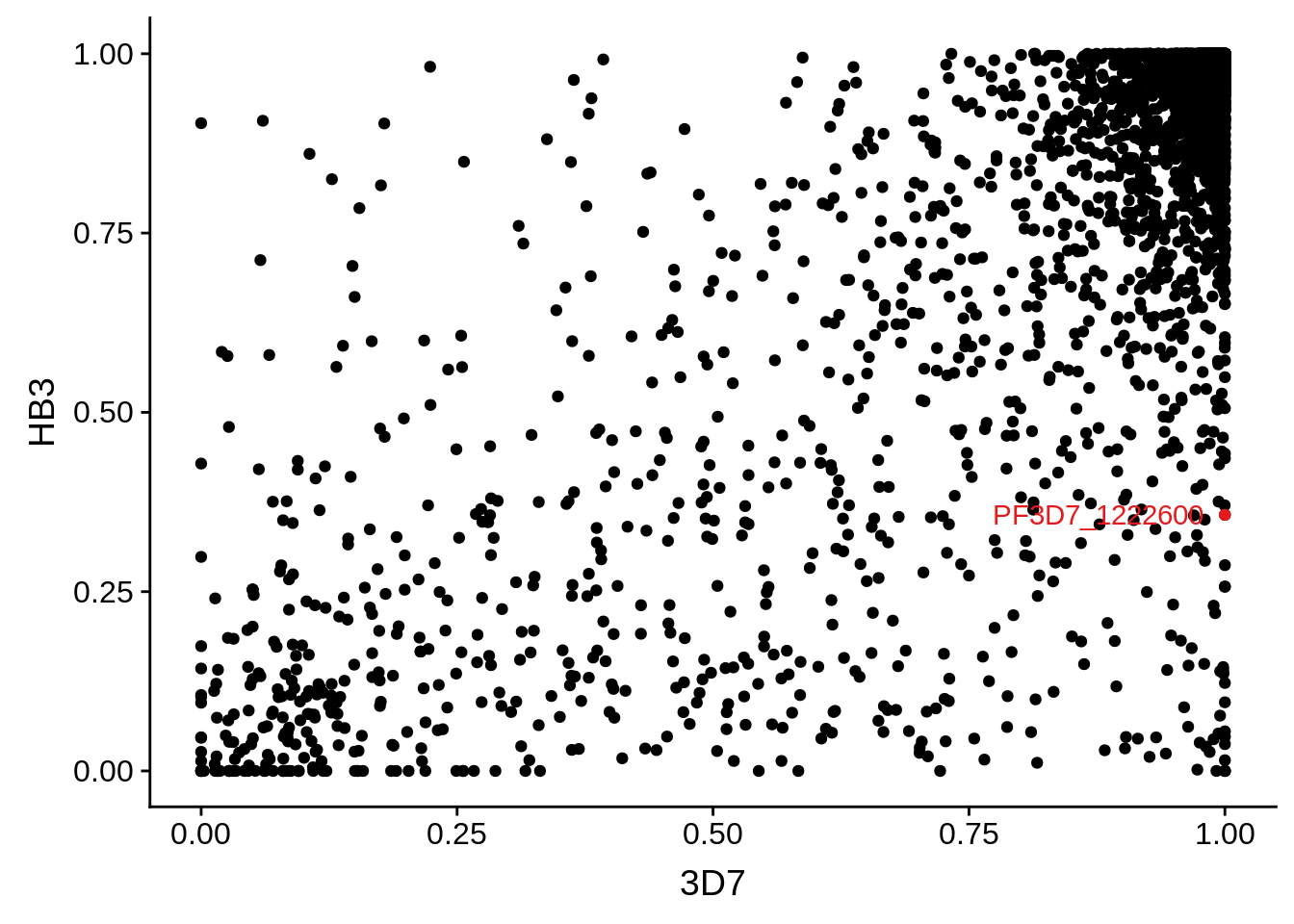
g <- ggplot(mean_coverages,aes(x=`3D7`,y=HB3)) + geom_point() + geom_point(data=mean_coverages %>% dplyr::filter(gene_id == "PF3D7_0930300"),color = "#E41A1C") + geom_text(data=mean_coverages %>% dplyr::filter(gene_id == "PF3D7_0930300"),label="PF3D7_0930300", color = "#E41A1C", hjust = 1.1)
print(g)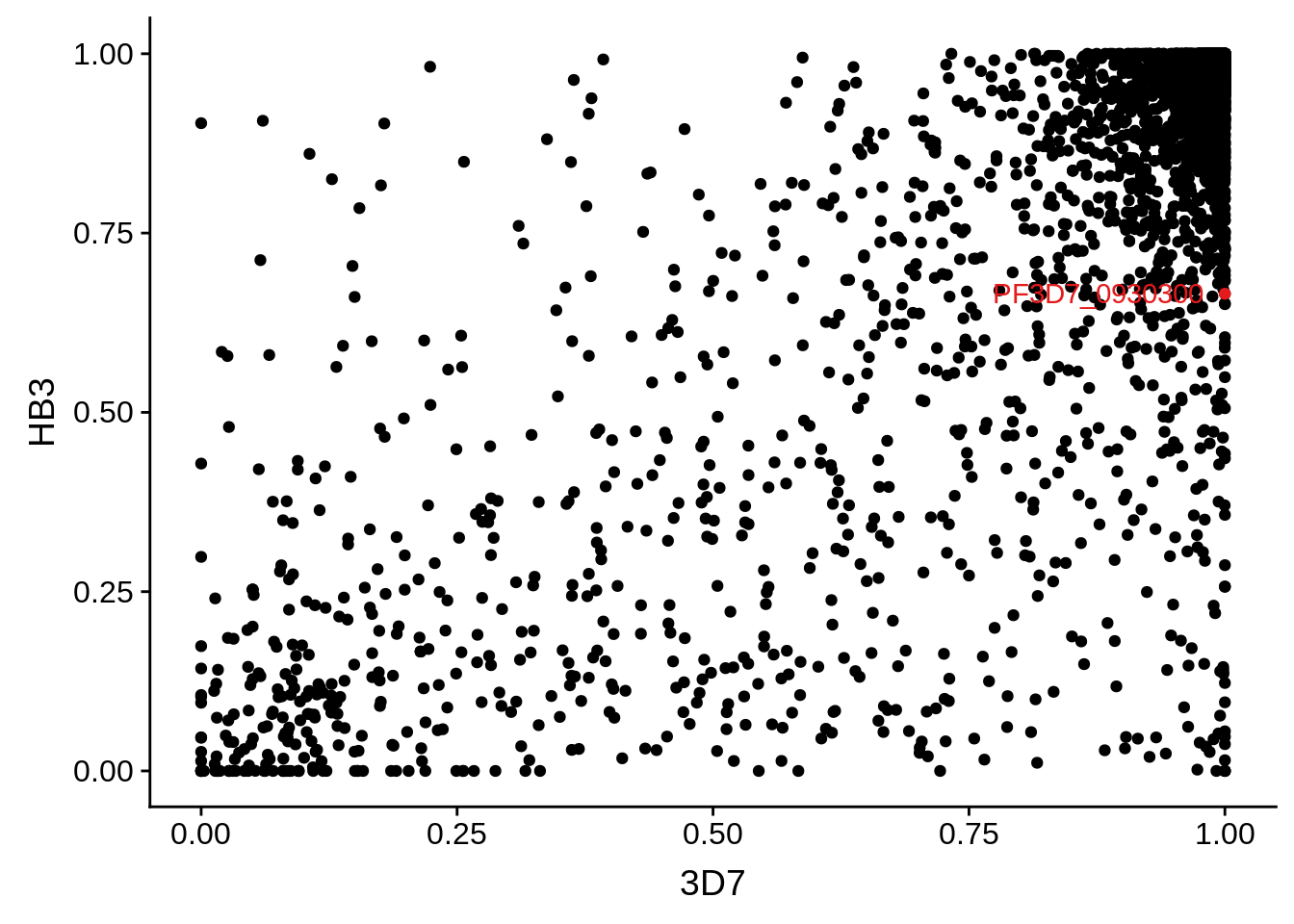
g <- ggplot(mean_coverages,aes(x=`3D7`,y=HB3)) + geom_point() + geom_point(data=mean_coverages %>% dplyr::filter(`3D7`<0.25 & HB3 >0.75),color = "#E41A1C") + geom_text(data=mean_coverages %>% dplyr::filter(`3D7`<0.25 & HB3 >0.75), aes(label=gene_id), color = "#E41A1C", hjust = -0.5, check_overlap = TRUE)
print(g)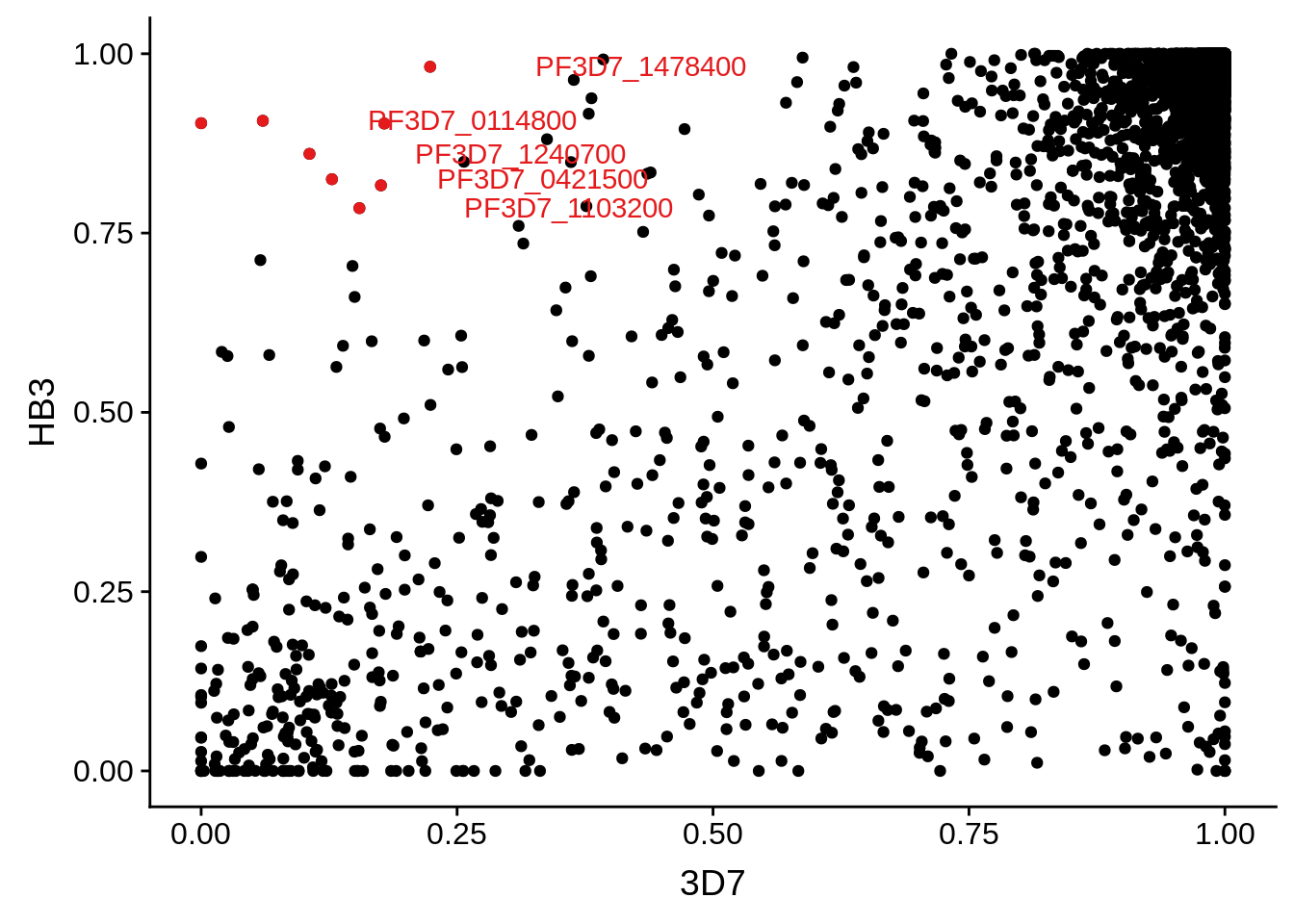
g <- ggplot(mean_coverages,aes(x=`3D7`,y=IT)) + geom_point() + geom_point(data=mean_coverages %>% dplyr::filter(`3D7`<0.5 & IT >0.5),color = "#E41A1C") + geom_text(data=mean_coverages %>% dplyr::filter(`3D7`<0.5 & IT >0.5), aes(label=gene_id), color = "#E41A1C", hjust = -0.5, check_overlap = TRUE)
print(g)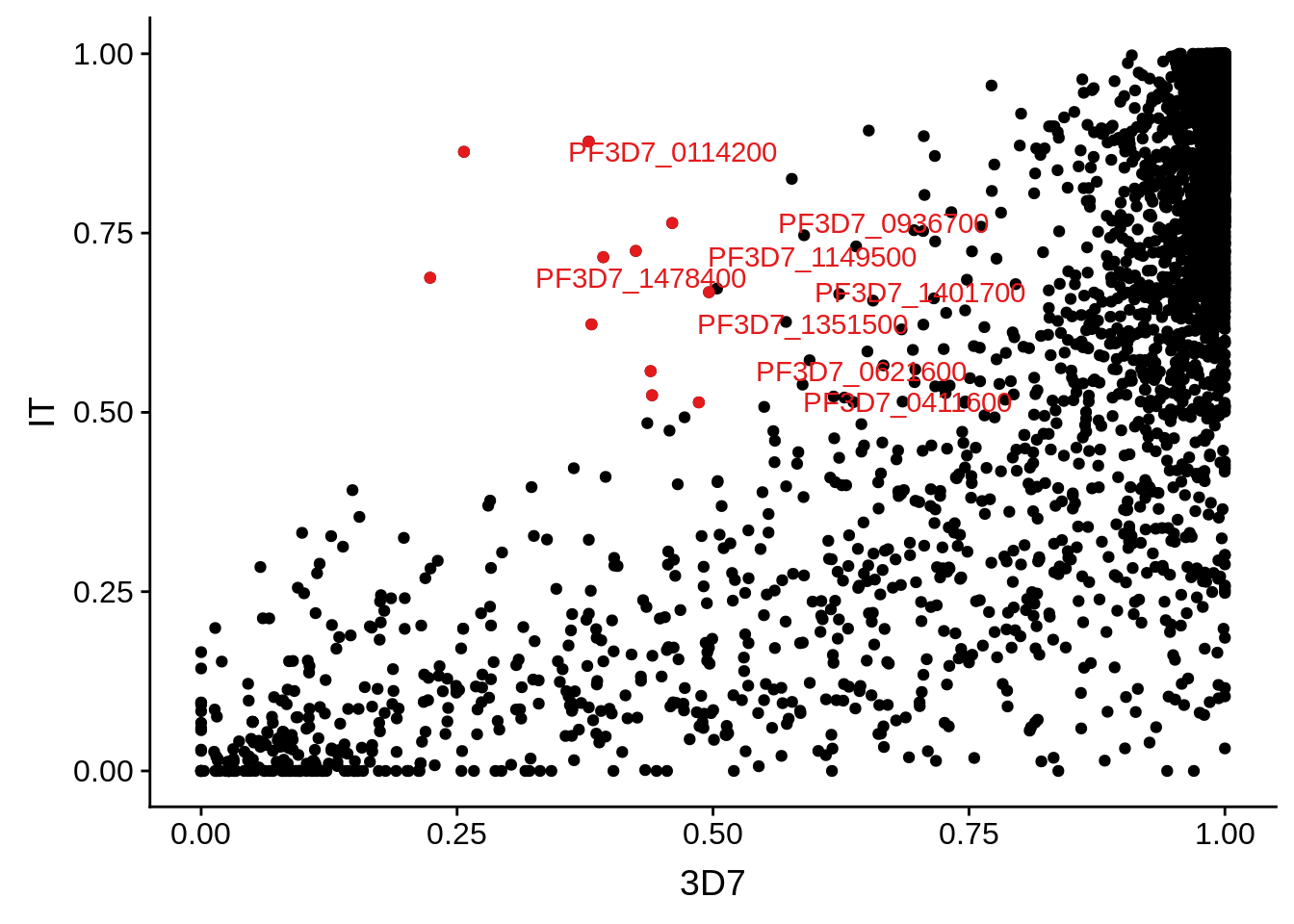
g <- ggplot(mean_coverages,aes(x=IT,y=HB3)) + geom_point() + geom_point(data=mean_coverages %>% dplyr::filter(IT<0.25 & HB3 >0.75),color = "#E41A1C") + geom_text(data=mean_coverages %>% dplyr::filter(IT<0.25 & HB3 >0.75), aes(label=gene_id), color = "#E41A1C", hjust = -0.5, check_overlap = TRUE)
print(g)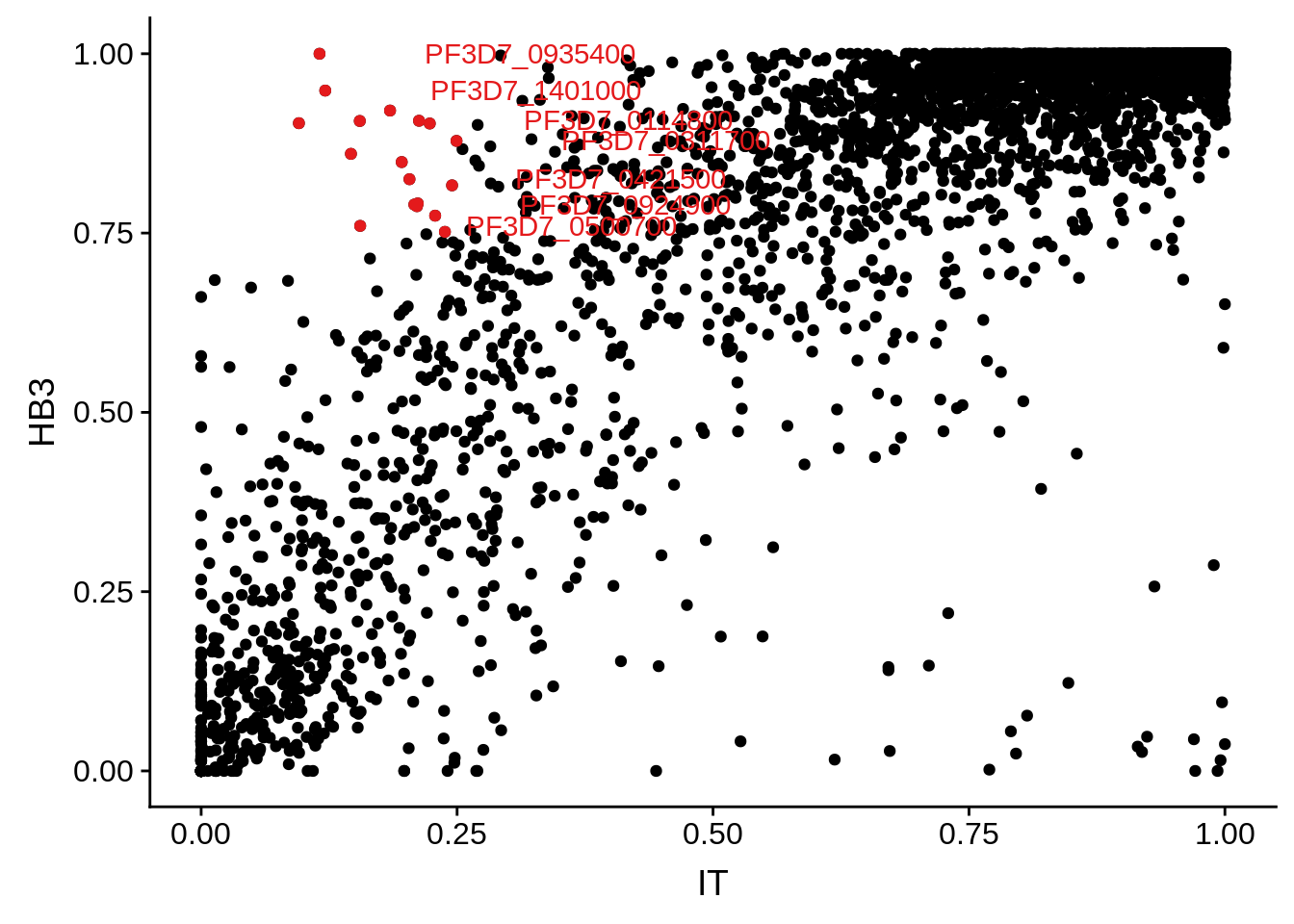
Results
Some genes may barely get over the threshold of 5 TPMs, which was chosen arbitrarily. We are not interested in those results. We want to see what transcripts are clearly abundant in one strain, while clearly absent or at the very least severely down-regulated in another strain. The question then becomes whether this is due to poor coverage across the gene (highly polymorphic) or actual low transcript copy number. MSP1 is an example of a highly polymorphic gene in HB3 versus 3D7. It is not covered very well across the gene and thus has been marked as “undetected.” MSP2 is an example of a gene that is detected, but is also highly polymorphic.
Looking through this CGH data will help as well: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2901668/
off_3d7_hb3 <- readr::read_tsv("../output/differential_detection/off_genes_3d7_hb3.tsv")
DT::datatable(off_3d7_hb3,rownames = F)off_3d7_it <- readr::read_tsv("../output/differential_detection/off_genes_3d7_hb3.tsv")
DT::datatable(off_3d7_hb3,rownames = F)off_hb3_it <- readr::read_tsv("../output/differential_detection/off_genes_3d7_hb3.tsv")
DT::datatable(off_3d7_hb3,rownames = F)Up in 3D7
PF3D7_0202000 2145.335 versus 2.995 knob-associated histidine-rich protein (KAHRP)
PF3D7_1372200 836.830 versus 0.599 histidine-rich protein III (HRPIII)
PF3D7_0201700 110.712 versus 0.052 DnaJ protein, putative
PF3D7_1429200 24.382 versus 0.99 transcription factor with AP2 domain(s)
PF3D7_0518700 30.287 versus 1.4 mRNA-binding protein PUF1 (PUF1)
PF3D7_1126800 37.441 versus 2.2023 RNA-binding protein, putative
PF3D7_1350900 43.551 versus 1.301 transcription factor with AP2 domain(s) (ApiAP2)
PF3D7_1222600 50.469 versus 0.951 transcription factor with AP2 domain(s) (AP2-G)
PF3D7_1241400 19.426 versus 1.633 RNA-binding protein, putative
PF3D7_0209000 34.424 versus 0.084 6-cysteine protein (P230)
PF3D7_1346700 46.211 versus 2.543 6-cysteine protein (P48/45)
PF3D7_1346800 13.754 versus 1.110 6-cysteine protein (P47)
PF3D7_1038400 14.784 versus 0.030 gametocyte-specific protein (Pf11-1)
PF3D7_0625600 31.212 versus 2.054 poly(A) polymerase PAP, putative
PF3D7_1031000 27.841 versus 0.468 25 kDa ookinete surface antigen precursor (Pfs25)
Up in HB3
10 out of top 30 are pseudogenes
Not many differences beyond highly variant genes (variant due to clonal variation or polymorphic regions? Both?)
stevor, exported protein family 4, pfmc-2tm mauler’s cleft two transmembrane protein, mspr4, g-beta repeat protein
3D7 and IT
Up in 3D7
See a lot of the same genes here as I did in 3D7 versus HB3.
PF3D7_1222600 transcription factor with AP2 domain(s) (AP2-G)
PF3D7_1346700 6-cysteine protein (P48/45)
PF3D7_0209000 6-cysteine protein (P230)
PF3D7_1477800 acyl-CoA binding protein (ACBP)
PF3D7_0414500 RNA-binding protein, putative
PF3D7_1031000 25 kDa ookinete surface antigen precursor (Pfs25)
PF3D7_1429200 transcription factor with AP2 domain(s)
PF3D7_0311700 plasmepsin VI
PF3D7_0109300 fatty acid elongation protein, GNS1/SUR4 family, putative
PF3D7_1038400 gametocyte-specific protein (Pf11-1)
####Up in IT
PF3D7_0114300 exported protein family 4, pseudogene (EPF4)
PF3D7_0936700 lysophospholipase, putative
HB3 and IT
Not much difference here.
Up in HB3
PF3D7_1477800 acyl-CoA binding protein (ACBP)
Up in IT
PF3D7_0202000 knob-associated histidine-rich protein (KAHRP)
PF3D7_1372200 histidine-rich protein III (HRPIII)
PF3D7_0201800 knob associated heat shock protein 40 (KAHsp40)
PF3D7_0201700 DnaJ protein, putative
PF3D7_0625600 poly(A) polymerase PAP, putative
Session Information
R version 3.5.0 (2018-04-23)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Gentoo/Linux
Matrix products: default
BLAS: /usr/local/lib64/R/lib/libRblas.so
LAPACK: /usr/local/lib64/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] bindrcpp_0.2.2
[2] BSgenome.Pfalciparum.PlasmoDB.v24_1.0
[3] BSgenome_1.48.0
[4] rtracklayer_1.40.6
[5] Biostrings_2.48.0
[6] XVector_0.20.0
[7] GenomicRanges_1.32.7
[8] GenomeInfoDb_1.16.0
[9] org.Pf.plasmo.db_3.6.0
[10] AnnotationDbi_1.42.1
[11] IRanges_2.14.12
[12] S4Vectors_0.18.3
[13] Biobase_2.40.0
[14] BiocGenerics_0.26.0
[15] scales_1.0.0
[16] cowplot_0.9.3
[17] magrittr_1.5
[18] forcats_0.3.0
[19] stringr_1.3.1
[20] dplyr_0.7.6
[21] purrr_0.2.5
[22] readr_1.1.1
[23] tidyr_0.8.1
[24] tibble_1.4.2
[25] ggplot2_3.0.0
[26] tidyverse_1.2.1
loaded via a namespace (and not attached):
[1] nlme_3.1-137 bitops_1.0-6
[3] matrixStats_0.54.0 lubridate_1.7.4
[5] bit64_0.9-7 httr_1.3.1
[7] rprojroot_1.3-2 tools_3.5.0
[9] backports_1.1.2 DT_0.4
[11] R6_2.3.0 DBI_1.0.0
[13] lazyeval_0.2.1 colorspace_1.3-2
[15] withr_2.1.2 tidyselect_0.2.4
[17] bit_1.1-14 compiler_3.5.0
[19] git2r_0.23.0 cli_1.0.1
[21] rvest_0.3.2 xml2_1.2.0
[23] DelayedArray_0.6.6 labeling_0.3
[25] digest_0.6.17 Rsamtools_1.32.3
[27] rmarkdown_1.10 R.utils_2.7.0
[29] pkgconfig_2.0.2 htmltools_0.3.6
[31] htmlwidgets_1.3 rlang_0.2.2
[33] readxl_1.1.0 rstudioapi_0.8
[35] RSQLite_2.1.1 shiny_1.1.0
[37] bindr_0.1.1 jsonlite_1.5
[39] crosstalk_1.0.0 BiocParallel_1.14.2
[41] R.oo_1.22.0 RCurl_1.95-4.11
[43] GenomeInfoDbData_1.1.0 Matrix_1.2-14
[45] Rcpp_0.12.19 munsell_0.5.0
[47] R.methodsS3_1.7.1 stringi_1.2.4
[49] yaml_2.2.0 SummarizedExperiment_1.10.1
[51] zlibbioc_1.26.0 plyr_1.8.4
[53] grid_3.5.0 blob_1.1.1
[55] promises_1.0.1 crayon_1.3.4
[57] lattice_0.20-35 haven_1.1.2
[59] hms_0.4.2 knitr_1.20
[61] pillar_1.3.0 XML_3.98-1.16
[63] glue_1.3.0 evaluate_0.11
[65] modelr_0.1.2 httpuv_1.4.5
[67] cellranger_1.1.0 gtable_0.2.0
[69] assertthat_0.2.0 mime_0.5
[71] xtable_1.8-3 broom_0.5.0
[73] later_0.7.5 GenomicAlignments_1.16.0
[75] memoise_1.1.0 workflowr_1.1.1 This R Markdown site was created with workflowr