Comparing predicted UTRs
Philipp Ross
2018-09-24
Last updated: 2018-09-24
Code version: f0b9974
library(org.Pf.plasmo.db)
aliases <- tibble::as_tibble(data.frame(org.Pf.plasmoALIAS2ORF))
utrs <- tibble::as_tibble(rtracklayer::import.gff("../output/final_utrs/final_5utrs_3d7.gff"))
utrs$Parent <- unlist(utrs$Parent)# Comparing our UTR estimates to the Derisi predictions
derisi1 <- tibble::as_tibble(rtracklayer::import.bed("../data/compare_utrs/GSM1410291_UTRs_1.bed"))
derisi2 <- tibble::as_tibble(rtracklayer::import.bed("../data/compare_utrs/GSM1410292_UTRs_2.bed"))
derisi3 <- tibble::as_tibble(rtracklayer::import.bed("../data/compare_utrs/GSM1410293_UTRs_3.bed"))
derisi4 <- tibble::as_tibble(rtracklayer::import.bed("../data/compare_utrs/GSM1410294_UTRs_4.bed"))
derisi5 <- tibble::as_tibble(rtracklayer::import.bed("../data/compare_utrs/GSM1410295_UTRs_5.bed"))
fix_derisi_utrs <- function(set) {
set$name <- stringi::stri_replace_last(set$name,replacement=" ",regex="_")
set <- set %>% tidyr::separate(name,into = c("gene_id","type"), sep =" ")
set$gene_id <- toupper(set$gene_id)
out <- dplyr::inner_join(set, aliases, by=c("gene_id"="alias_symbol"))
return(out)
}
derisi1 <- fix_derisi_utrs(derisi1)
derisi2 <- fix_derisi_utrs(derisi2)
derisi3 <- fix_derisi_utrs(derisi3)
derisi4 <- fix_derisi_utrs(derisi4)
derisi5 <- fix_derisi_utrs(derisi5)tmp1 <- dplyr::select(derisi1, gene_id.y, width)
tmp2 <- dplyr::select(utrs, Parent, width)
compare_derisi <- dplyr::inner_join(tmp1,tmp2,by=c("gene_id.y"="Parent"))
rm(tmp1,tmp2)
ggplot(compare_derisi,aes(width.x,width.y)) + geom_point() + geom_smooth(method="lm")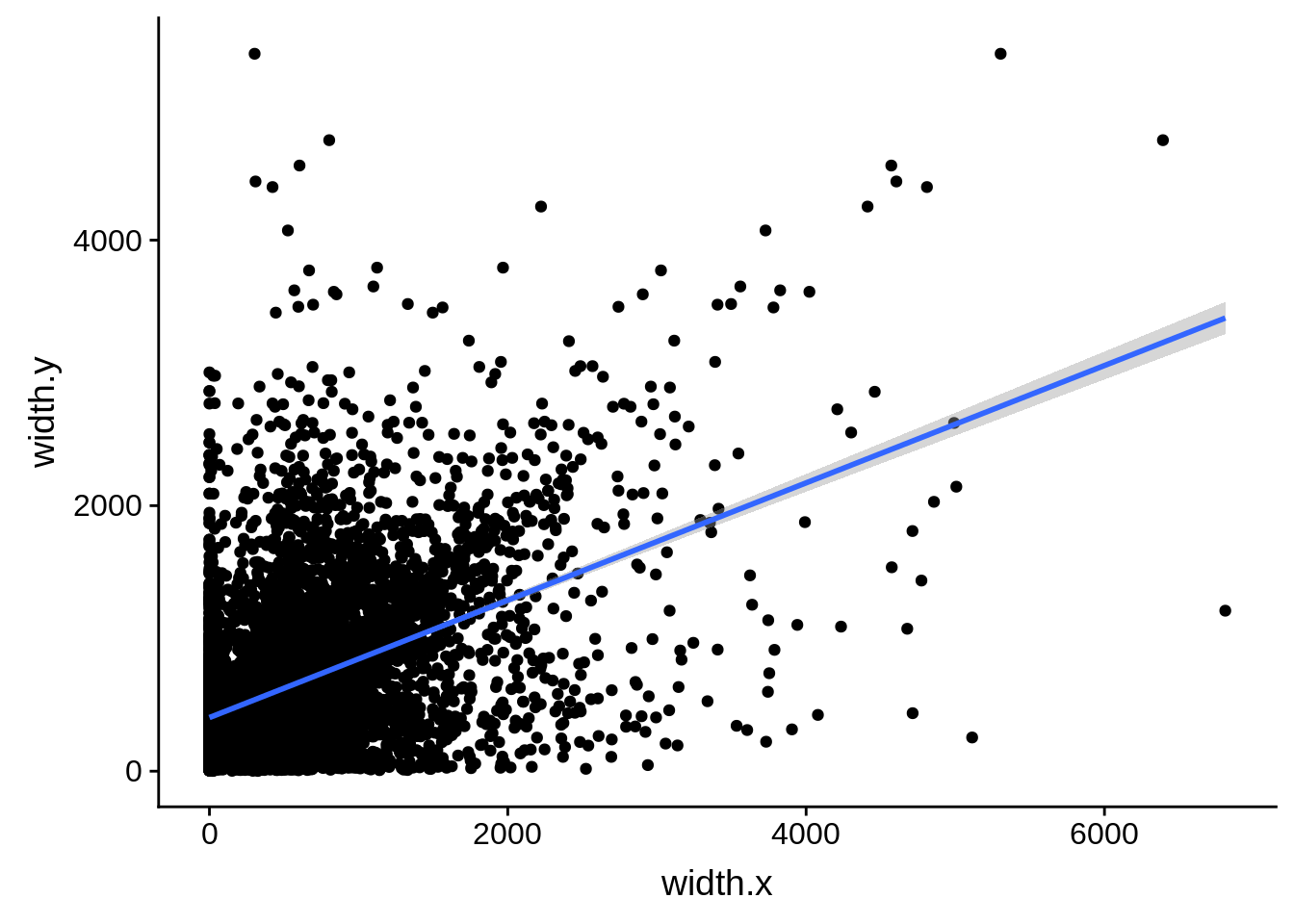
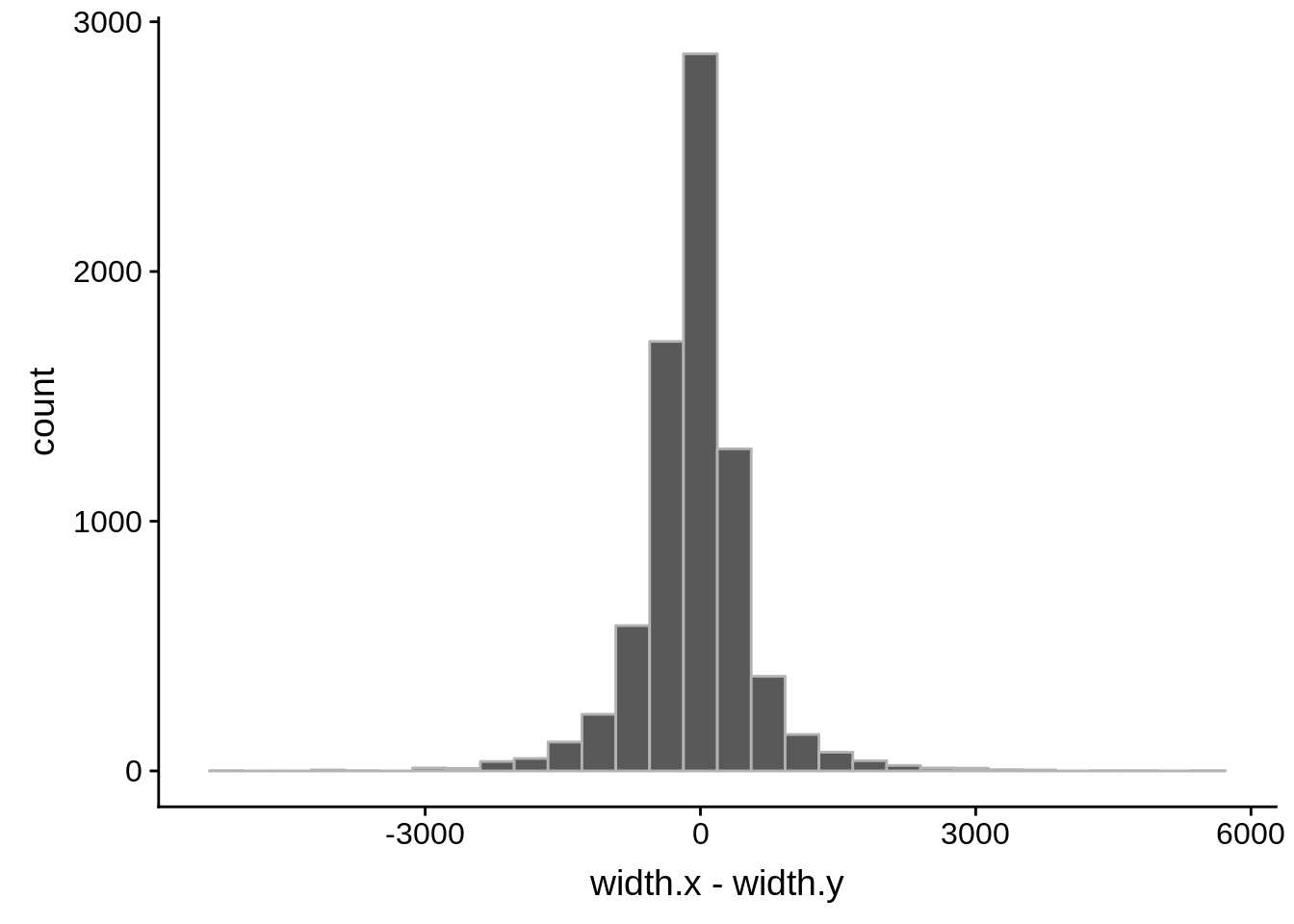
tmp1 <- dplyr::select(derisi2, gene_id.y, width)
tmp2 <- dplyr::select(utrs, Parent, width)
compare_derisi <- dplyr::inner_join(tmp1,tmp2,by=c("gene_id.y"="Parent"))
rm(tmp1,tmp2)
ggplot(compare_derisi,aes(width.x,width.y)) + geom_point() + geom_smooth(method="lm")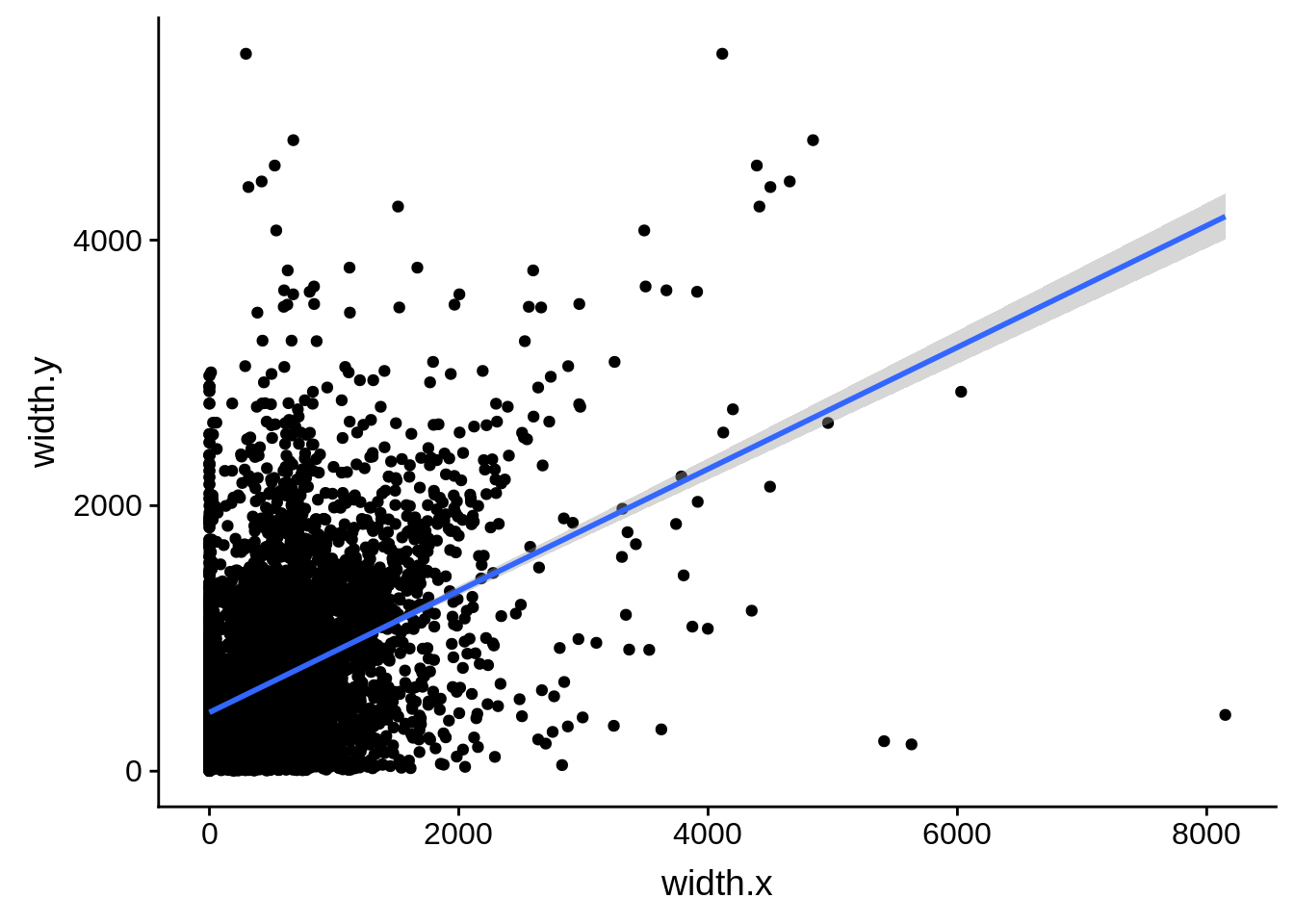
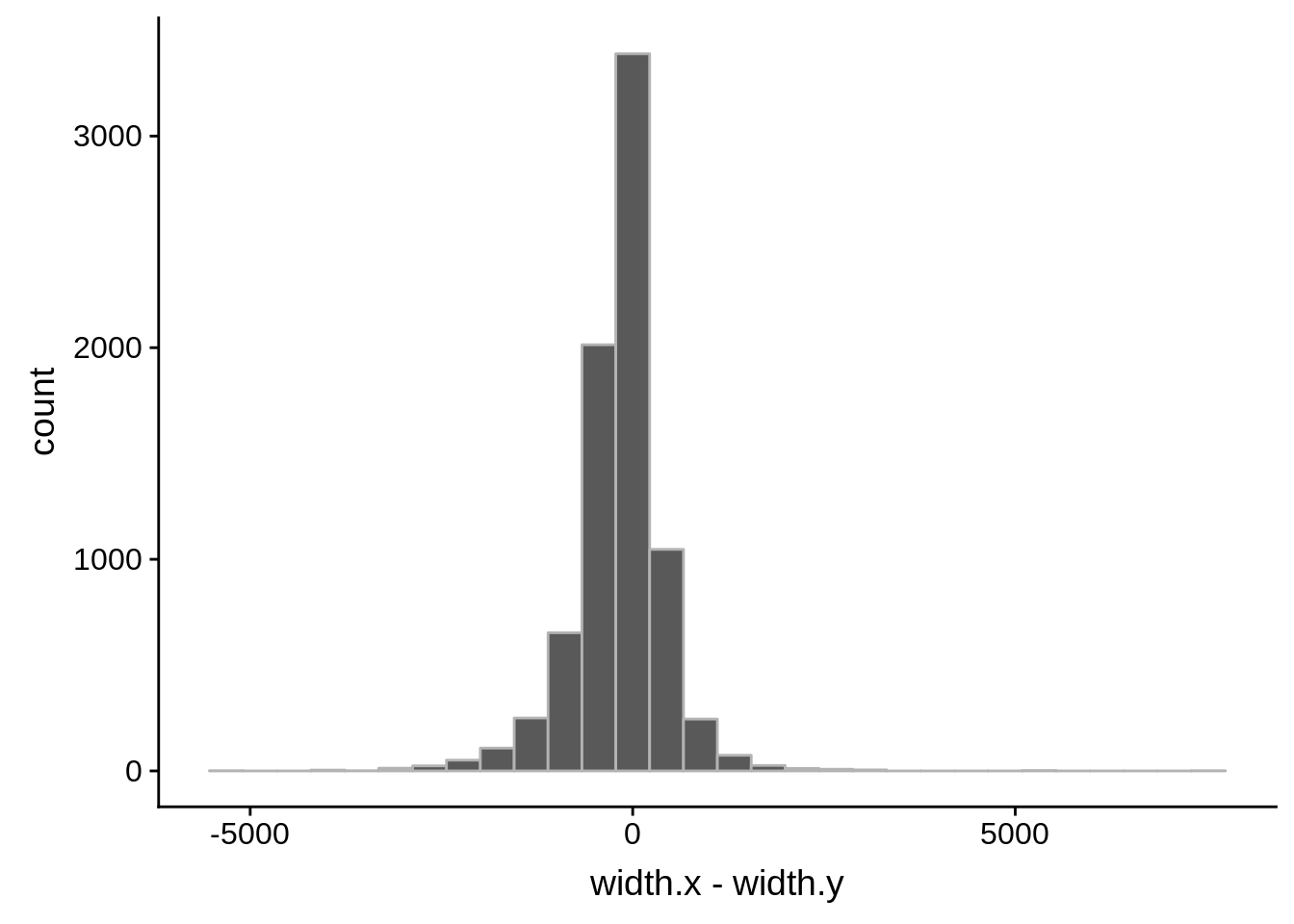
tmp1 <- dplyr::select(derisi3, gene_id.y, width)
tmp2 <- dplyr::select(utrs, Parent, width)
compare_derisi <- dplyr::inner_join(tmp1,tmp2,by=c("gene_id.y"="Parent"))
rm(tmp1,tmp2)
ggplot(compare_derisi,aes(width.x,width.y)) + geom_point() + geom_smooth(method="lm")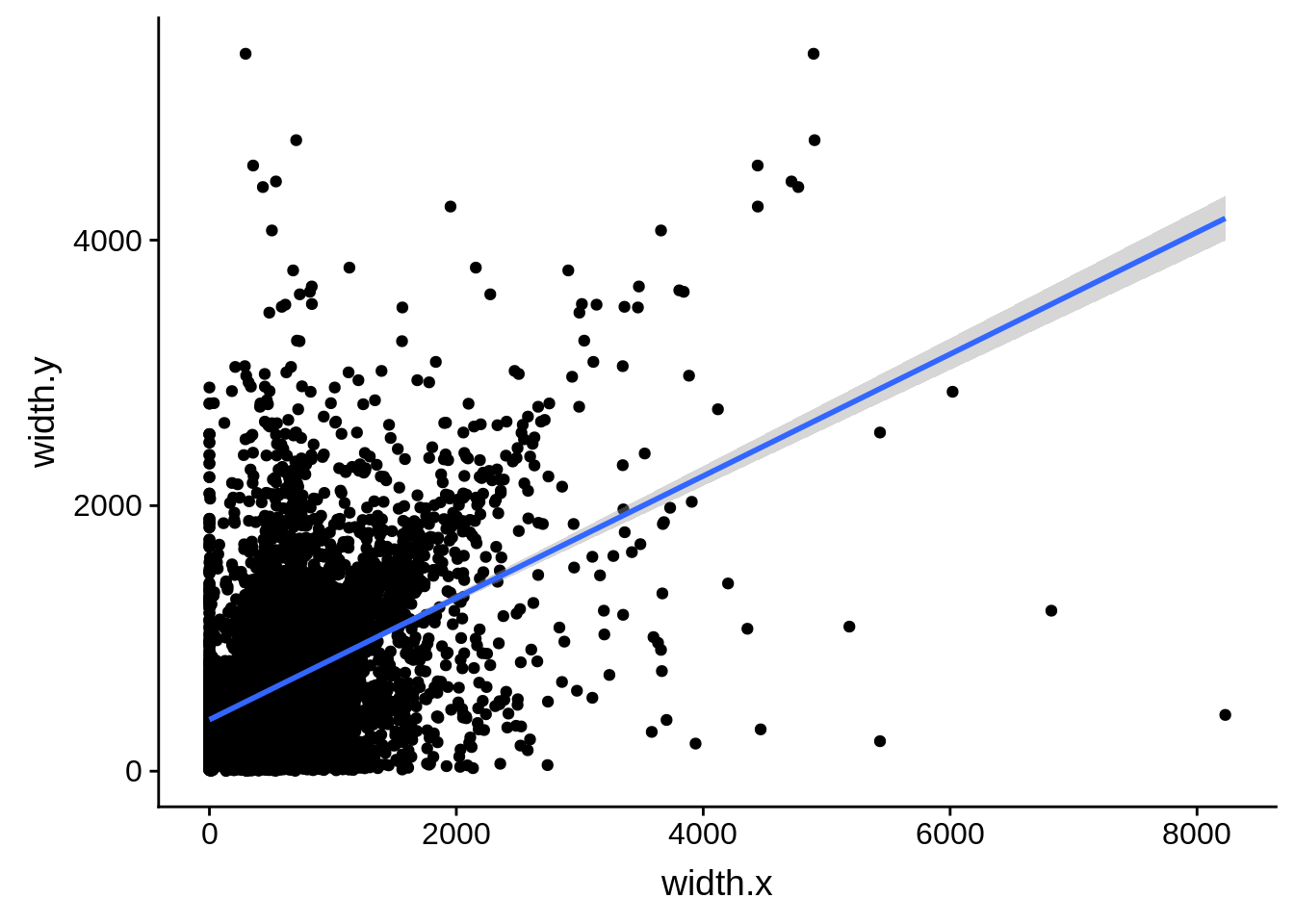
tmp1 <- dplyr::select(derisi3, gene_id.y, width)
tmp2 <- dplyr::select(utrs, Parent, width)
compare_derisi <- dplyr::inner_join(tmp1,tmp2,by=c("gene_id.y"="Parent"))
rm(tmp1,tmp2)
ggplot(compare_derisi,aes(width.x,width.y)) + geom_point() + geom_smooth(method="lm")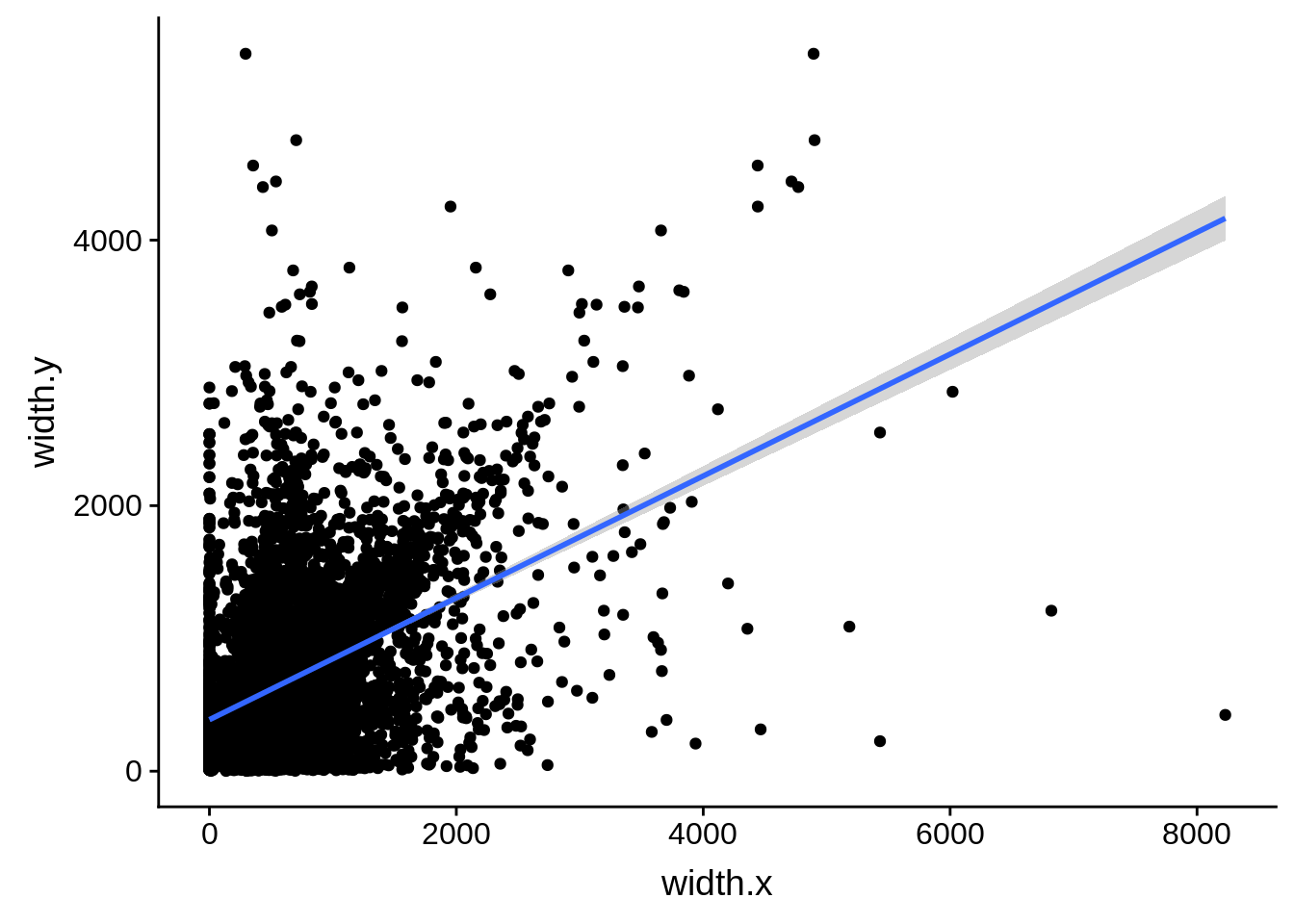
tmp1 <- dplyr::select(derisi4, gene_id.y, width)
tmp2 <- dplyr::select(utrs, Parent, width)
compare_derisi <- dplyr::inner_join(tmp1,tmp2,by=c("gene_id.y"="Parent"))
rm(tmp1,tmp2)
ggplot(compare_derisi,aes(width.x,width.y)) + geom_point() + geom_smooth(method="lm")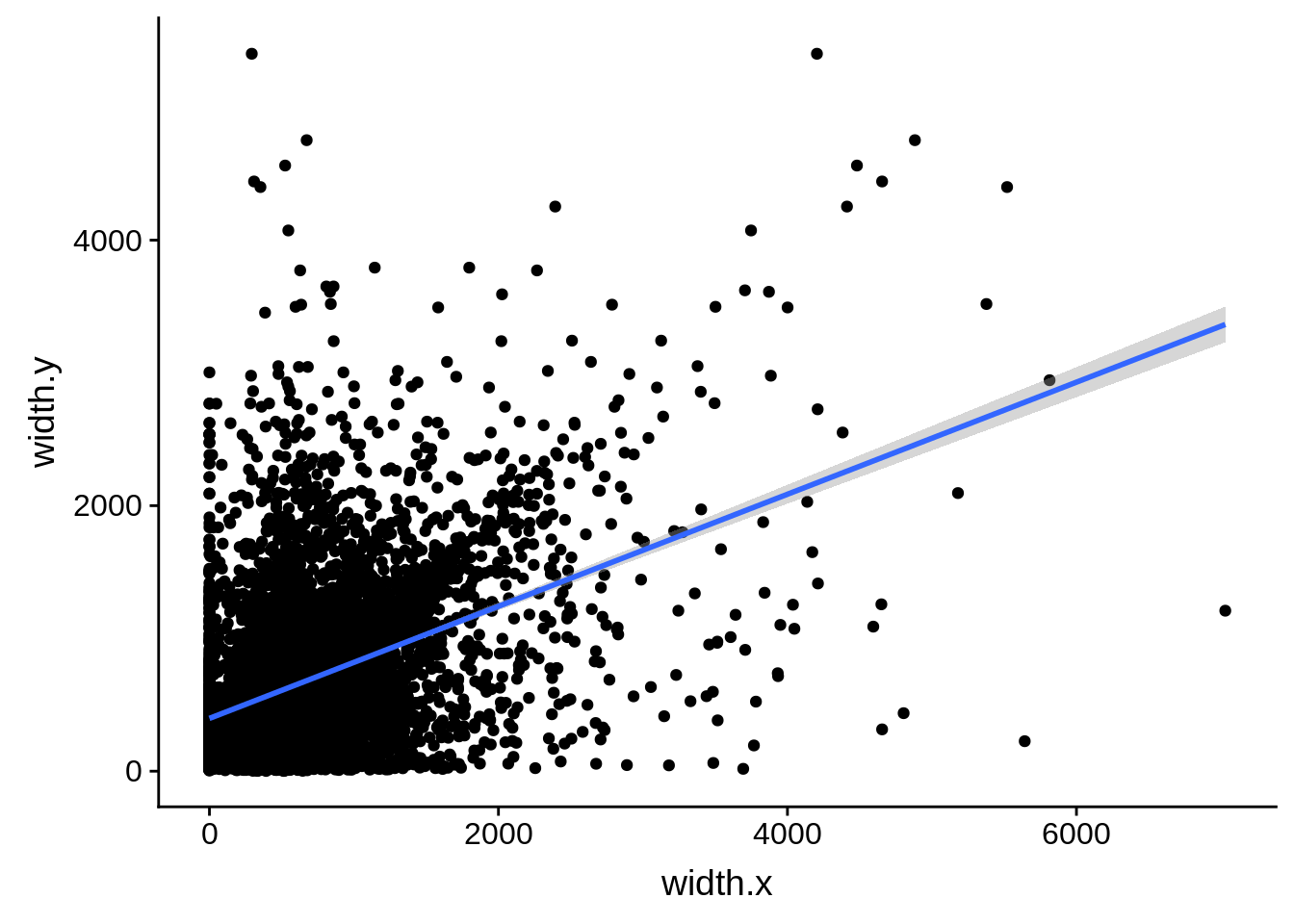
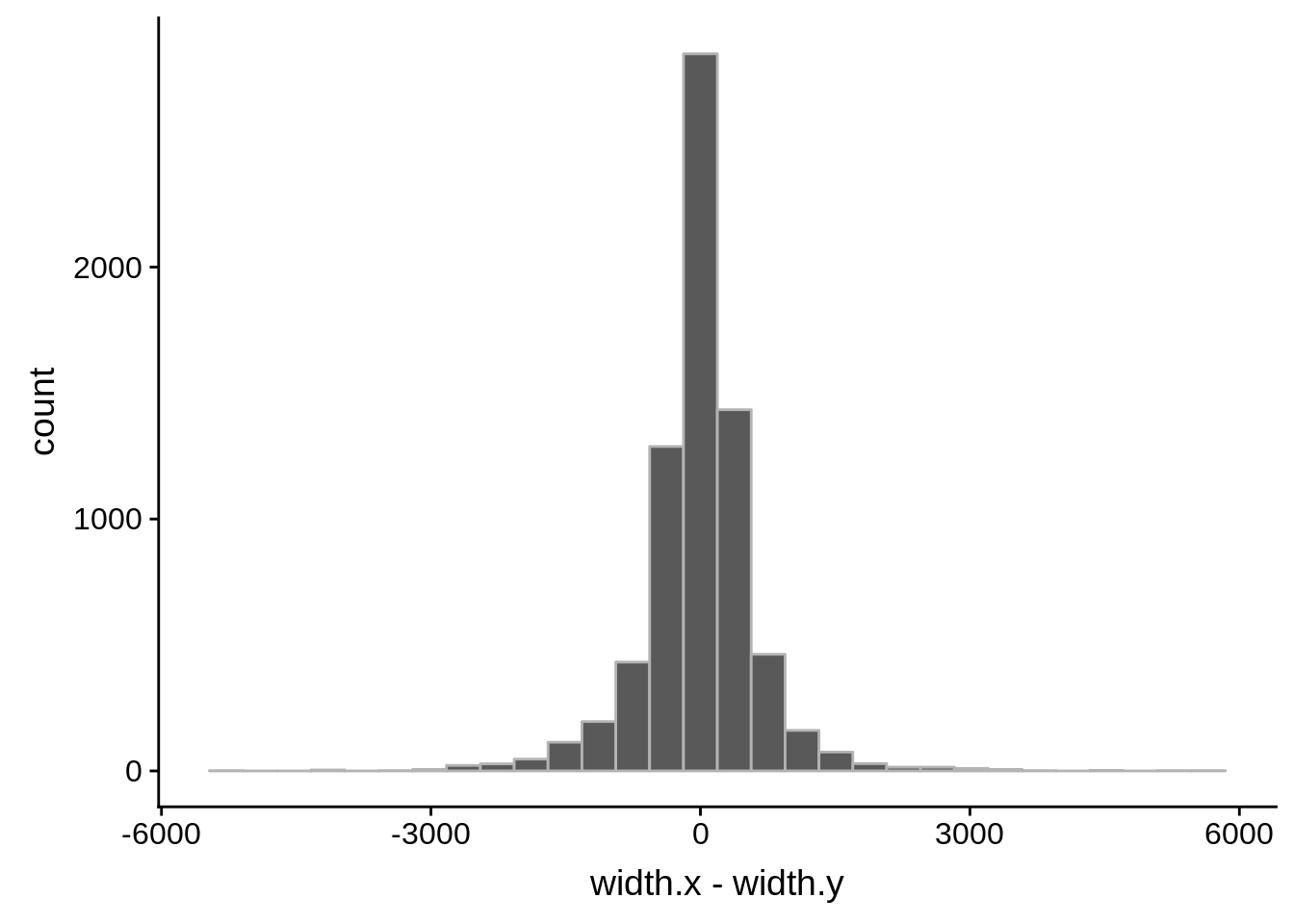
tmp1 <- dplyr::select(derisi5, gene_id.y, width)
tmp2 <- dplyr::select(utrs, Parent, width)
compare_derisi <- dplyr::inner_join(tmp1,tmp2,by=c("gene_id.y"="Parent"))
rm(tmp1,tmp2)
ggplot(compare_derisi,aes(width.x,width.y)) + geom_point() + geom_smooth(method="lm")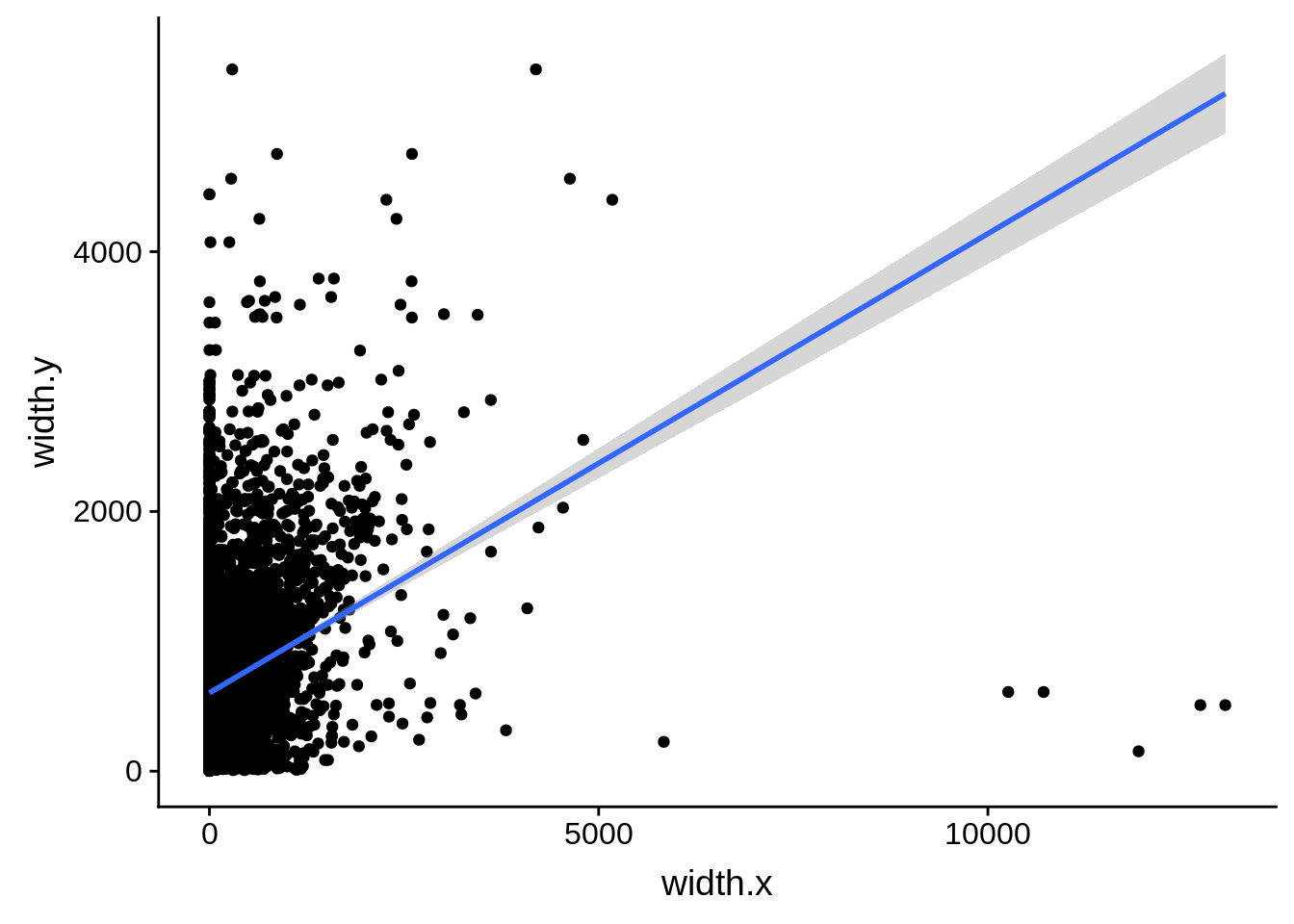
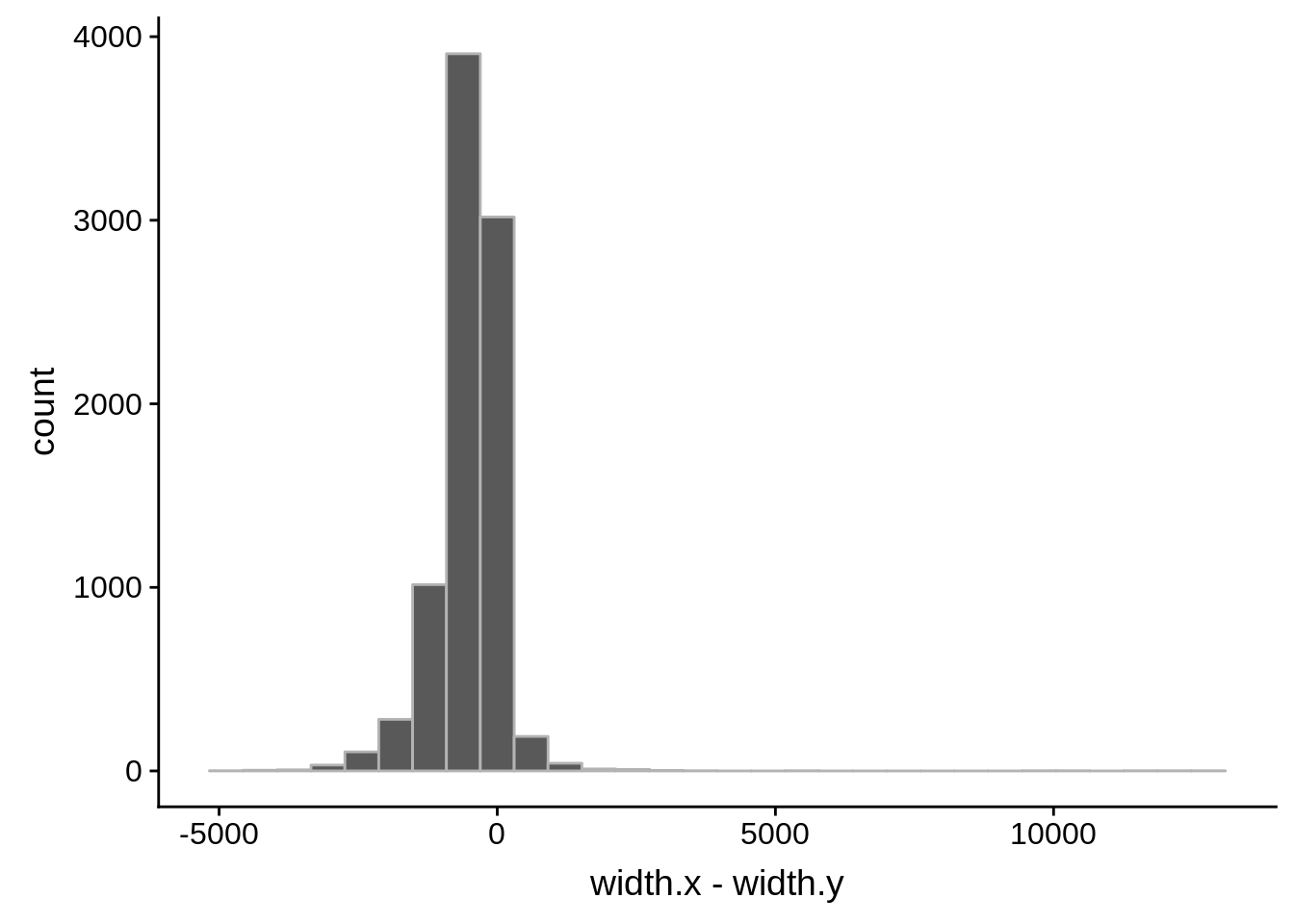
# Comparing our UTR estimates to the Adjalley predictions
adjalley <- tibble::as_tibble(rtracklayer::import.gff("../data/compare_utrs/sorted_Adjalley_Chabbert_TSSs.gff"))
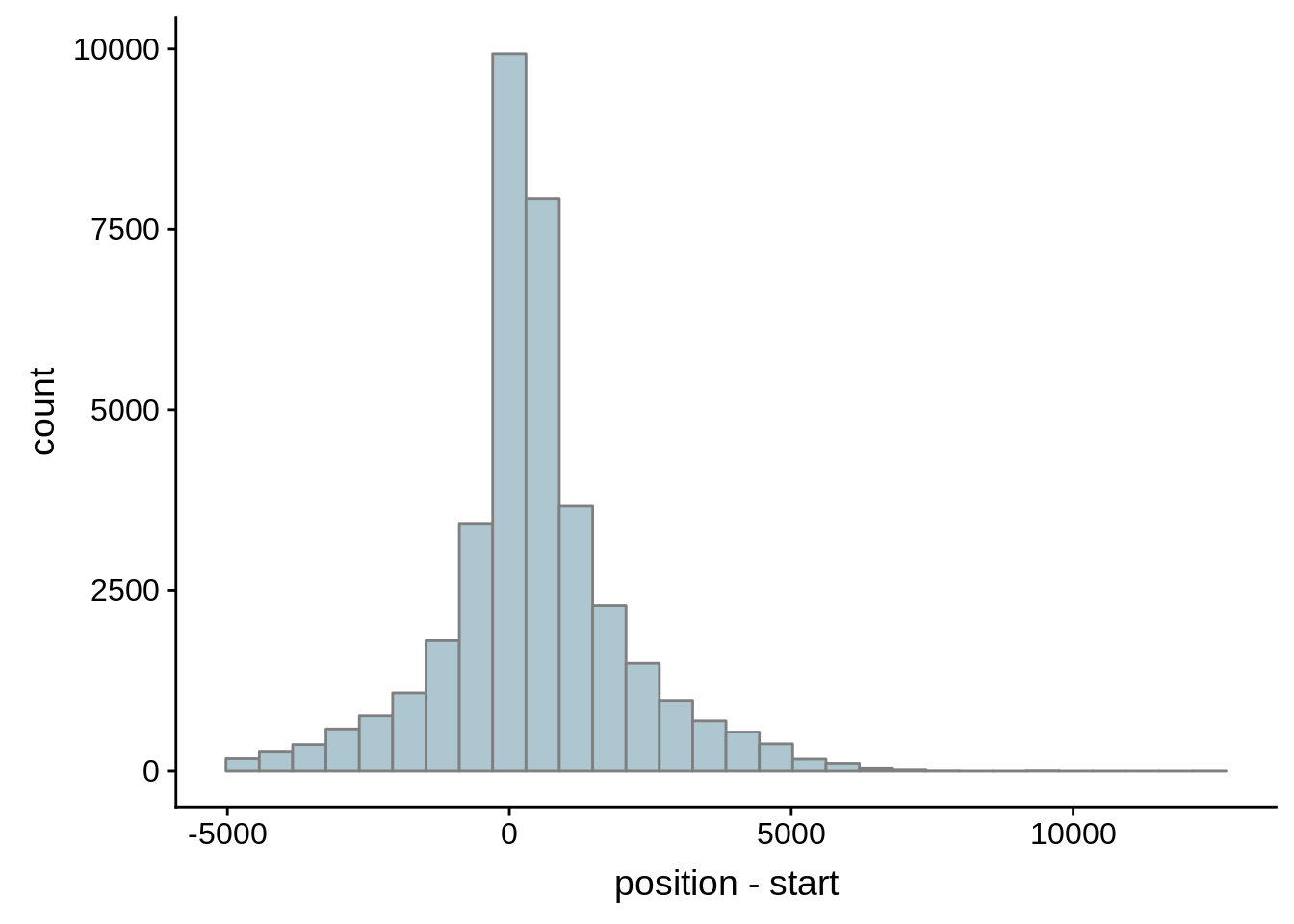
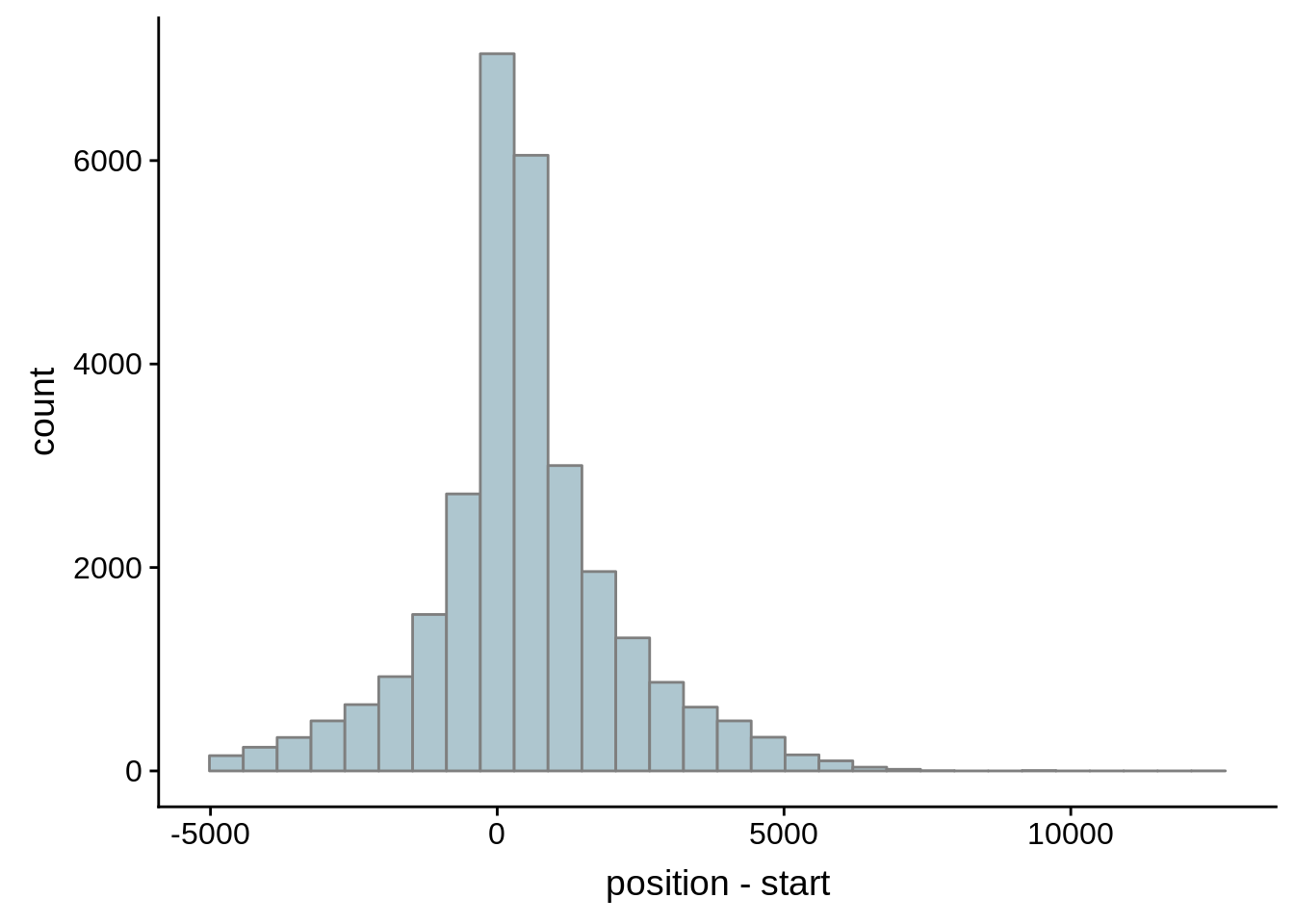
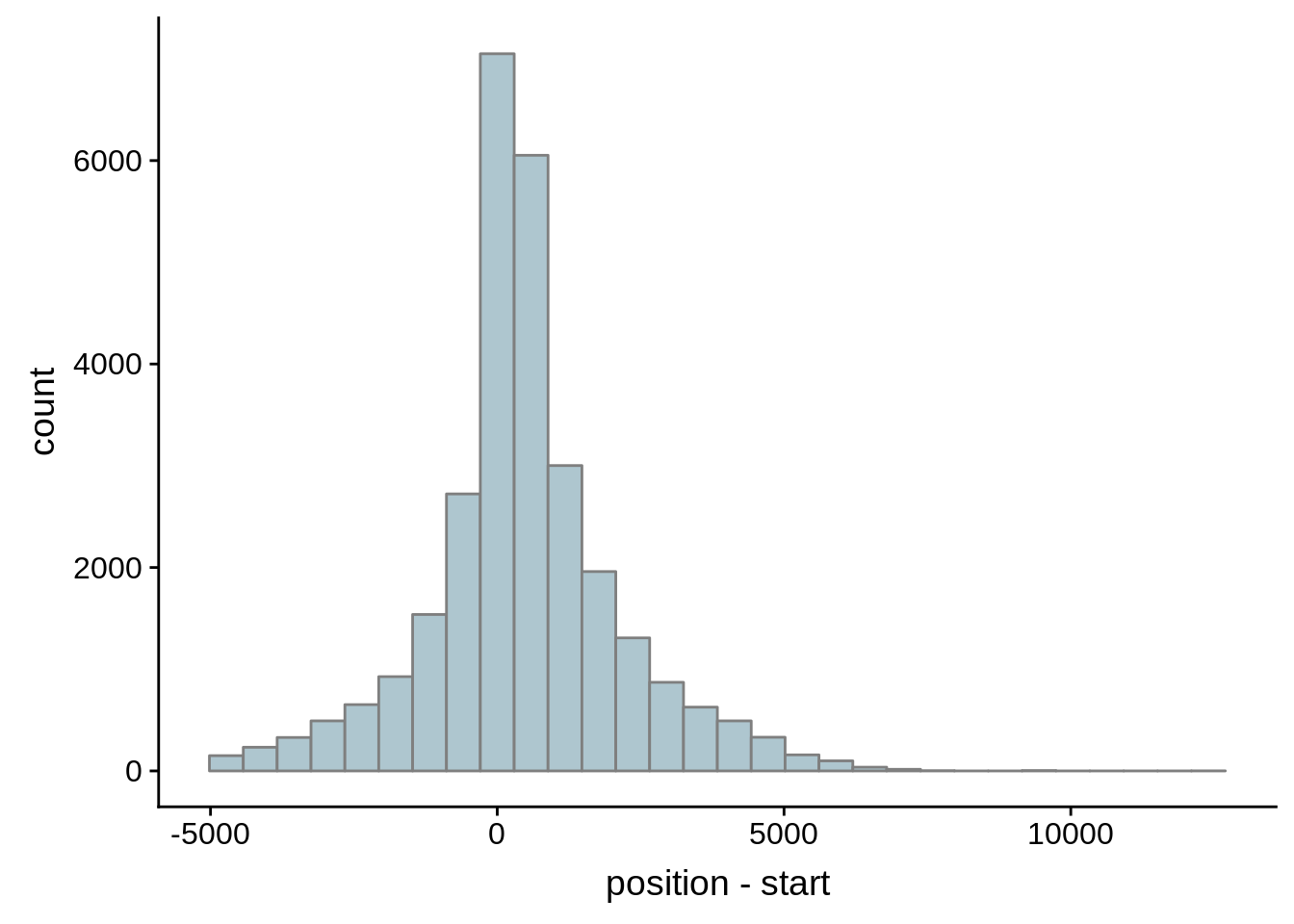
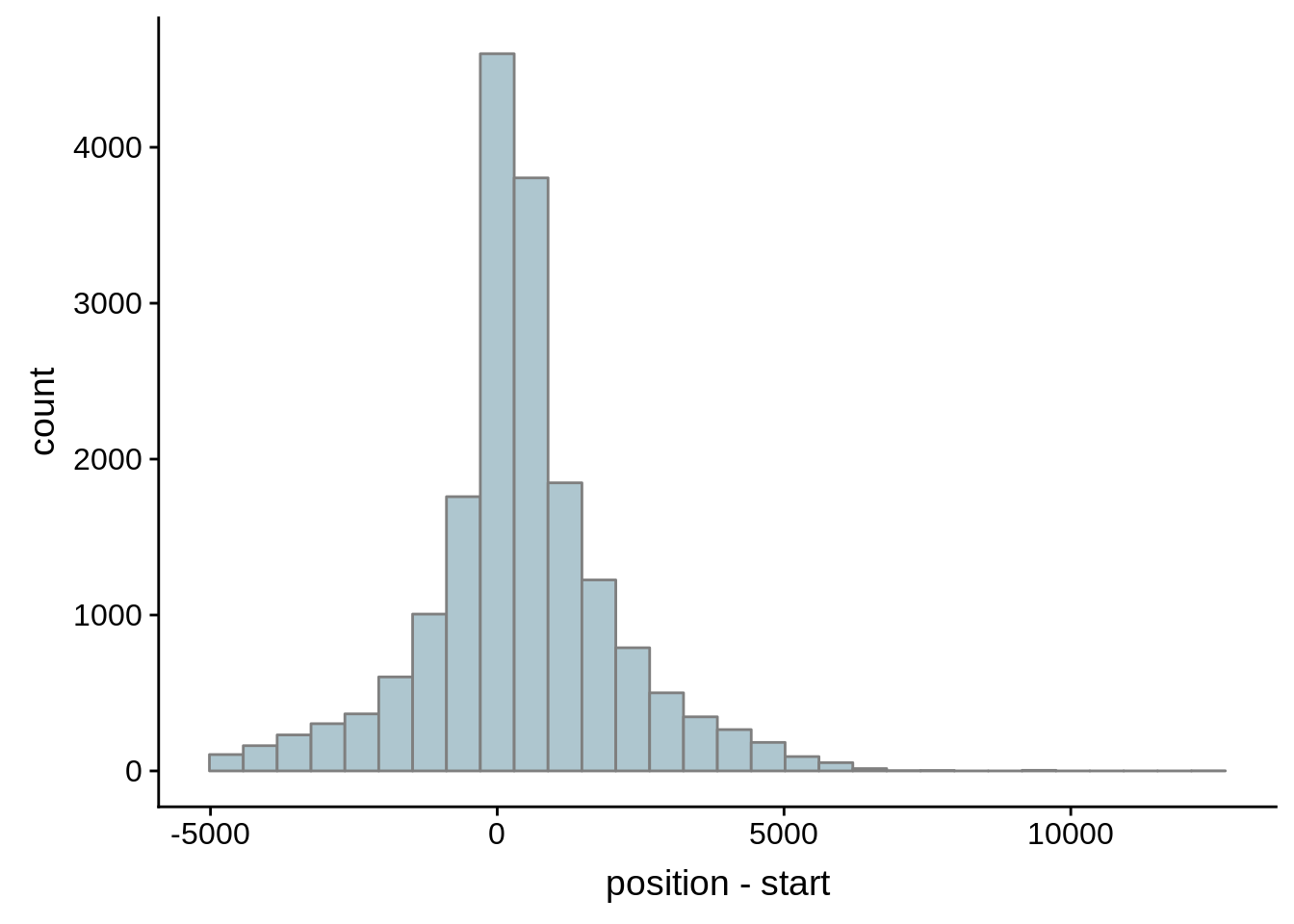
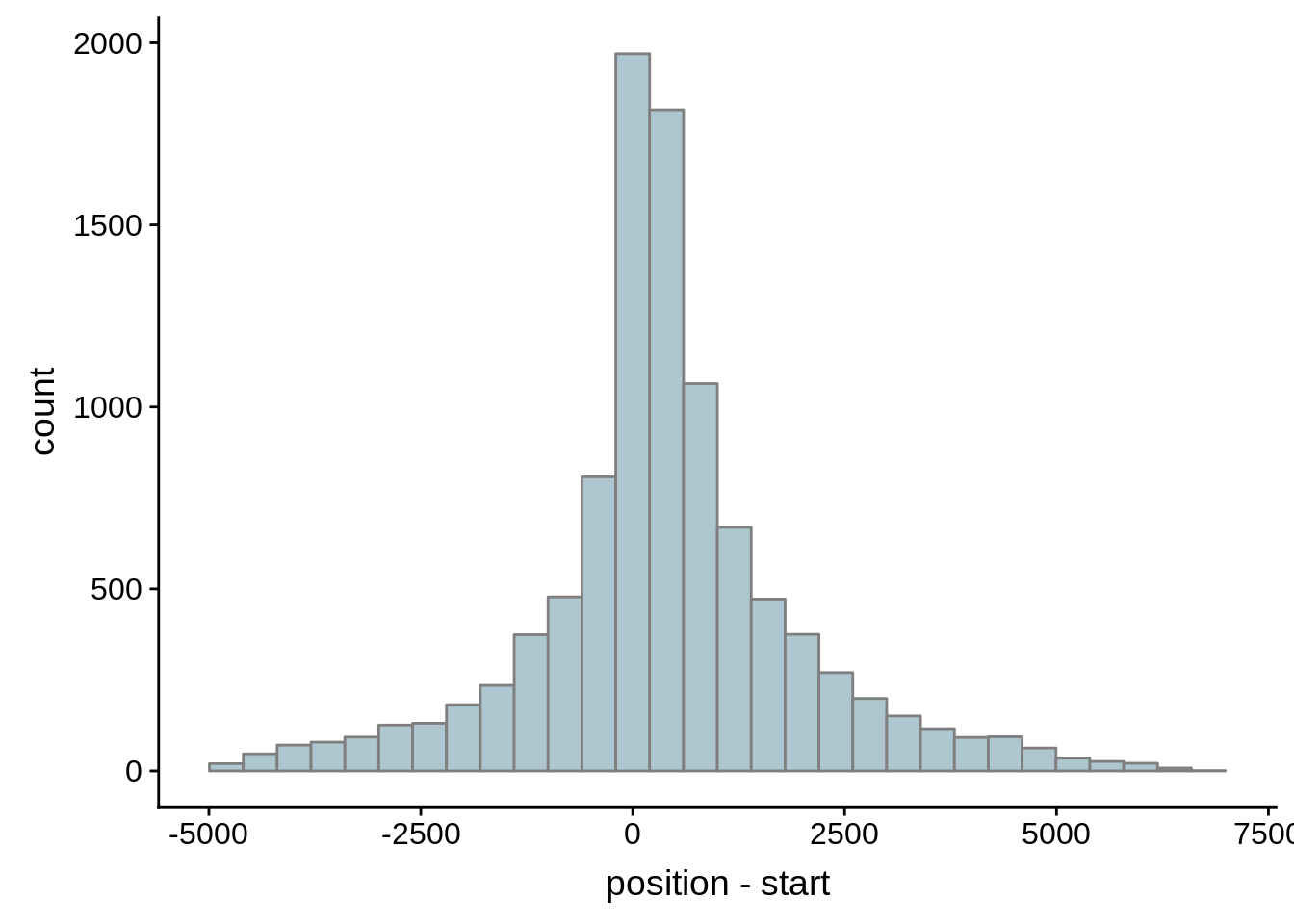
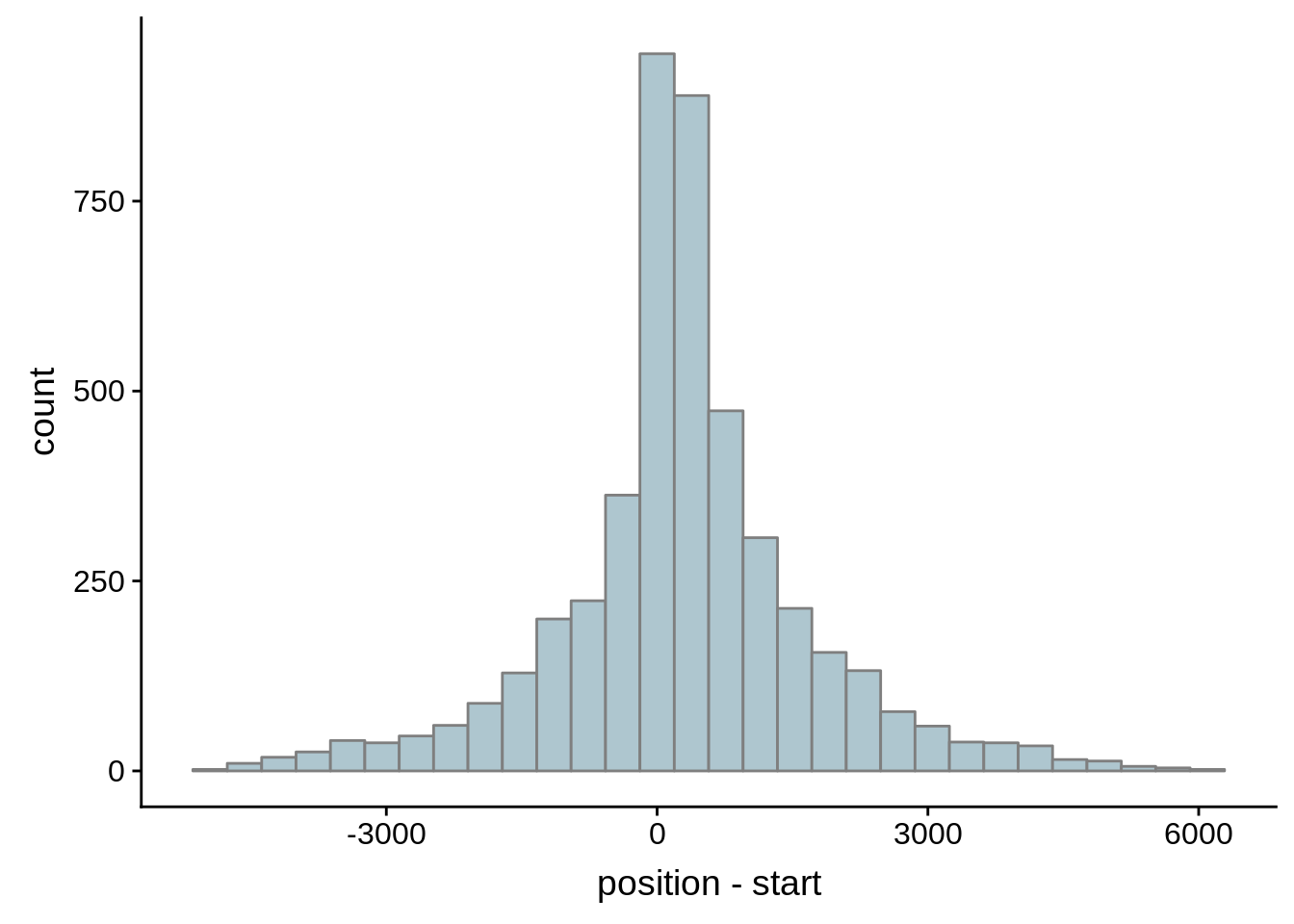
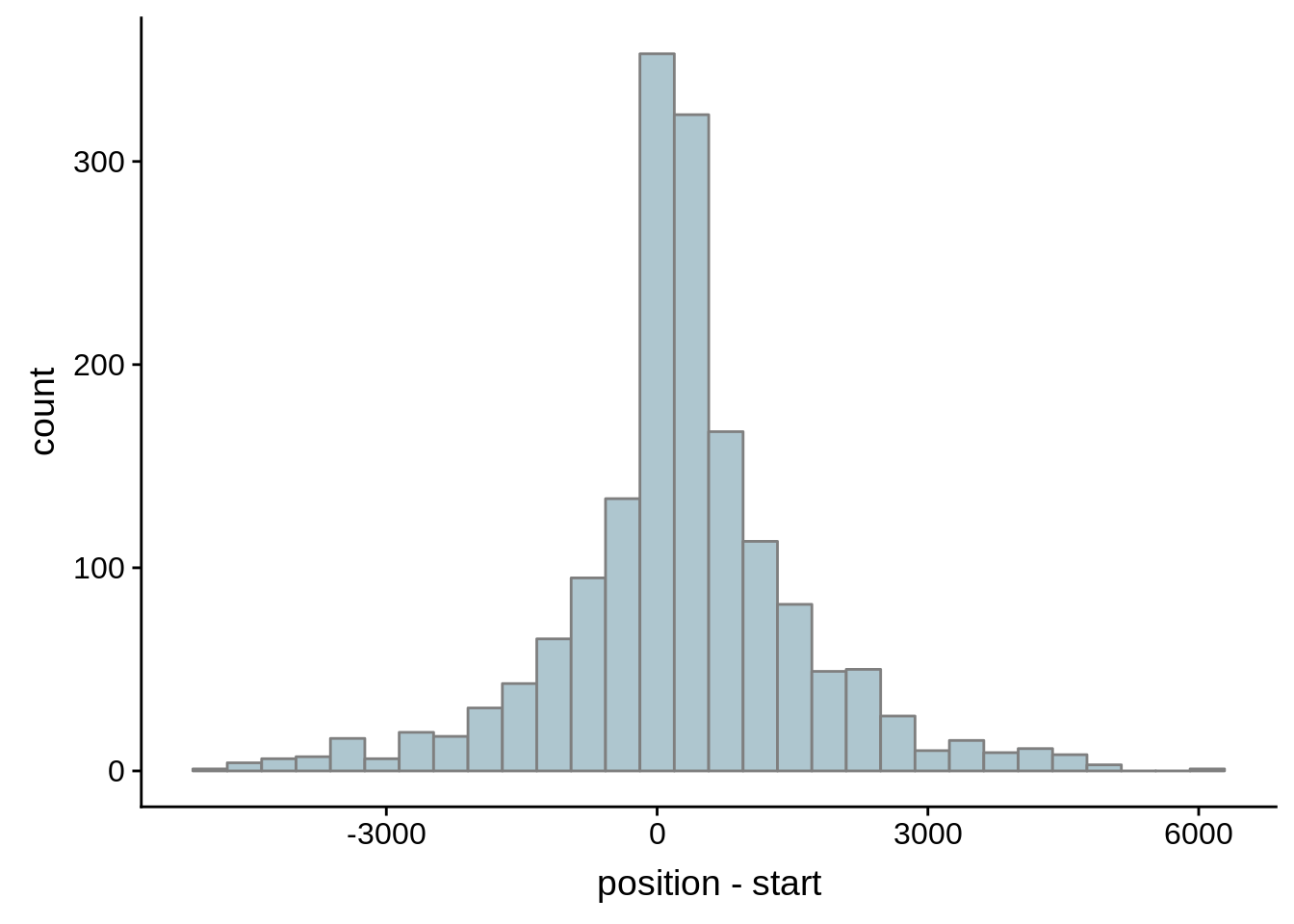
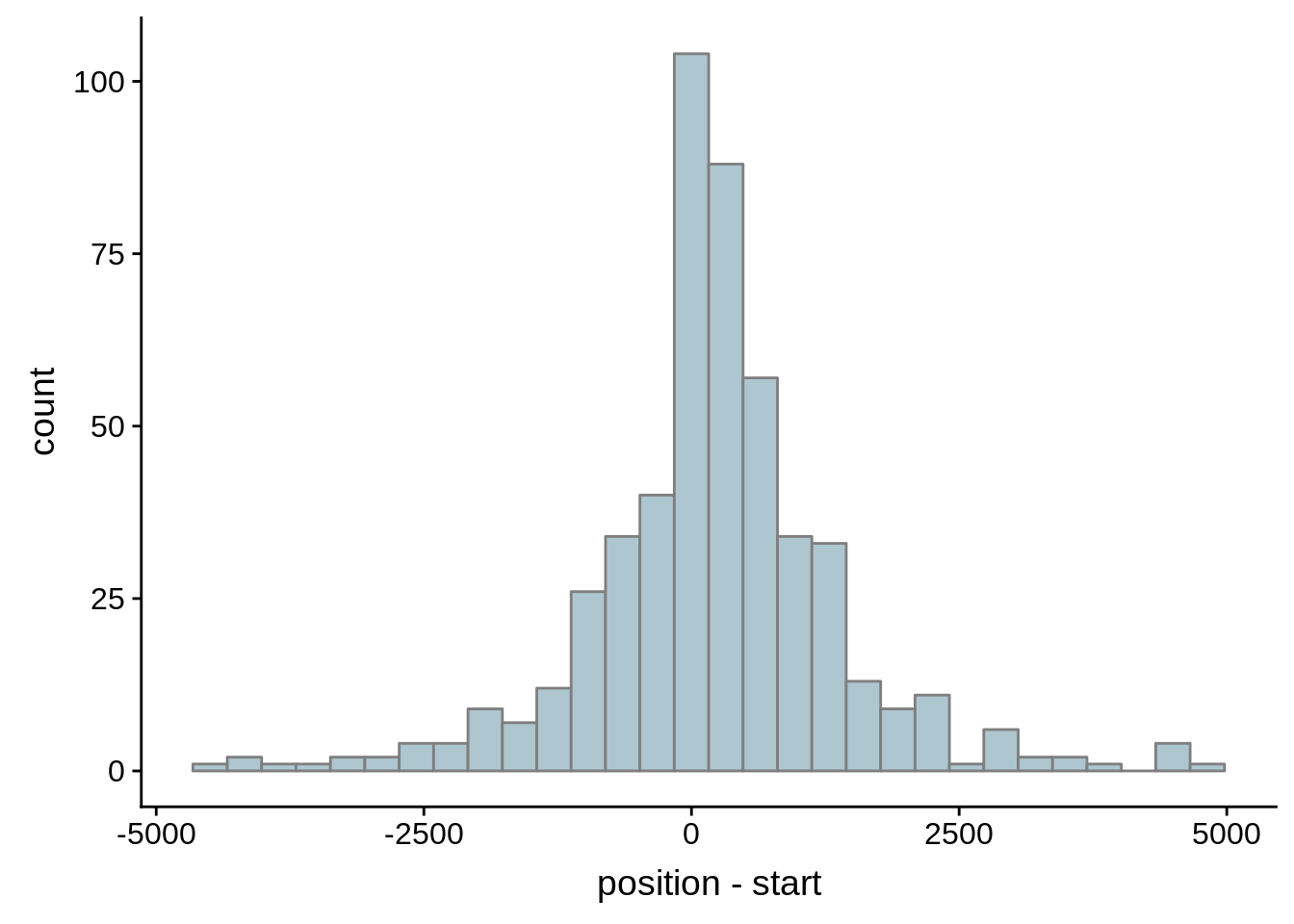
for (i in 1:8) {
adjalley_filtered <- adjalley %>%
dplyr::mutate(position=(start+end)/2) %>%
dplyr::filter(FilterSize>i)
tmp1 <- dplyr::select(adjalley_filtered, AssignedFeat, position)
tmp2 <- dplyr::select(utrs, Parent, start)
compare_adjalley <- dplyr::inner_join(tmp1,tmp2,by=c("AssignedFeat"="Parent"))
g <- ggplot(compare_adjalley,aes(position-start)) + geom_histogram(fill="#aec6cf",color="grey50")
print(g)
}
Session Information
R version 3.5.0 (2018-04-23)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Gentoo/Linux
Matrix products: default
BLAS: /usr/local/lib64/R/lib/libRblas.so
LAPACK: /usr/local/lib64/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] bindrcpp_0.2.2
[2] BSgenome.Pfalciparum.PlasmoDB.v24_1.0
[3] BSgenome_1.48.0
[4] rtracklayer_1.40.6
[5] Biostrings_2.48.0
[6] XVector_0.20.0
[7] GenomicRanges_1.32.6
[8] GenomeInfoDb_1.16.0
[9] org.Pf.plasmo.db_3.6.0
[10] AnnotationDbi_1.42.1
[11] IRanges_2.14.10
[12] S4Vectors_0.18.3
[13] Biobase_2.40.0
[14] BiocGenerics_0.26.0
[15] scales_1.0.0
[16] cowplot_0.9.3
[17] magrittr_1.5
[18] forcats_0.3.0
[19] stringr_1.3.1
[20] dplyr_0.7.6
[21] purrr_0.2.5
[22] readr_1.1.1
[23] tidyr_0.8.1
[24] tibble_1.4.2
[25] ggplot2_3.0.0
[26] tidyverse_1.2.1
loaded via a namespace (and not attached):
[1] nlme_3.1-137 bitops_1.0-6
[3] matrixStats_0.54.0 lubridate_1.7.4
[5] bit64_0.9-7 httr_1.3.1
[7] rprojroot_1.3-2 tools_3.5.0
[9] backports_1.1.2 R6_2.2.2
[11] DBI_1.0.0 lazyeval_0.2.1
[13] colorspace_1.3-2 withr_2.1.2
[15] tidyselect_0.2.4 bit_1.1-14
[17] compiler_3.5.0 git2r_0.23.0
[19] cli_1.0.0 rvest_0.3.2
[21] xml2_1.2.0 DelayedArray_0.6.5
[23] labeling_0.3 digest_0.6.15
[25] Rsamtools_1.32.3 rmarkdown_1.10
[27] R.utils_2.6.0 pkgconfig_2.0.2
[29] htmltools_0.3.6 rlang_0.2.2
[31] readxl_1.1.0 rstudioapi_0.7
[33] RSQLite_2.1.1 bindr_0.1.1
[35] jsonlite_1.5 BiocParallel_1.14.2
[37] R.oo_1.22.0 RCurl_1.95-4.11
[39] GenomeInfoDbData_1.1.0 Matrix_1.2-14
[41] Rcpp_0.12.18 munsell_0.5.0
[43] R.methodsS3_1.7.1 stringi_1.2.4
[45] yaml_2.2.0 SummarizedExperiment_1.10.1
[47] zlibbioc_1.26.0 plyr_1.8.4
[49] grid_3.5.0 blob_1.1.1
[51] crayon_1.3.4 lattice_0.20-35
[53] haven_1.1.2 hms_0.4.2
[55] knitr_1.20 pillar_1.3.0
[57] XML_3.98-1.16 glue_1.3.0
[59] evaluate_0.11 modelr_0.1.2
[61] cellranger_1.1.0 gtable_0.2.0
[63] assertthat_0.2.0 broom_0.5.0
[65] GenomicAlignments_1.16.0 memoise_1.1.0
[67] workflowr_1.1.1 This R Markdown site was created with workflowr