Intensity within and across plates
Joyce Hsiao
Last updated: 2017-12-11
Code version: ab4fe7a
Introduction/summary
Intensity distribution within each individual: for all three channels, we observe variation between single cell samples from the six individuals in both mean and variance; further, within each individual, the differences between single cell samples from different plates are dramatic, in both mean and variance.
Intensity correlation for each individual: high correlation between DAPI and GFP but not as much for between DAPI and RFP or between RFP and GREEN
Trying out PCA: The first PC explains 63% of variation in the data, and the second PC explains 27% of variation in the data. First PC correlated with RFP, GFP and DAPI and also correlated with nucleus size. The direction of correlation is the same for size and intensity metrics. Also, first PC is not correlated with batch, individual labels, perimeter or eccentricity.
Load data
\(~\)
ints <- readRDS(file="/project2/gilad/joycehsiao/fucci-seq/data/intensity.rds")Individual variation within plate and across plates
RFP
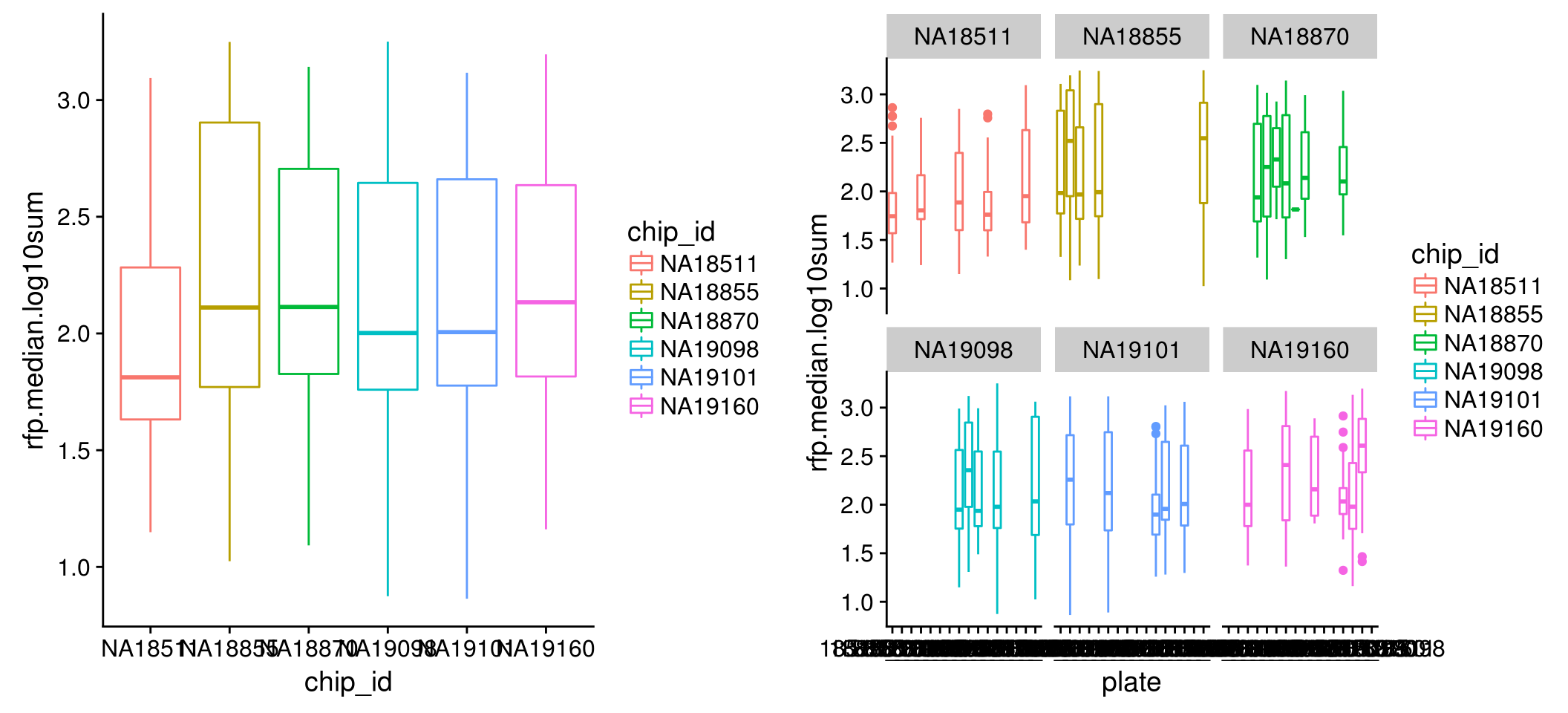
\(~\)
GFP
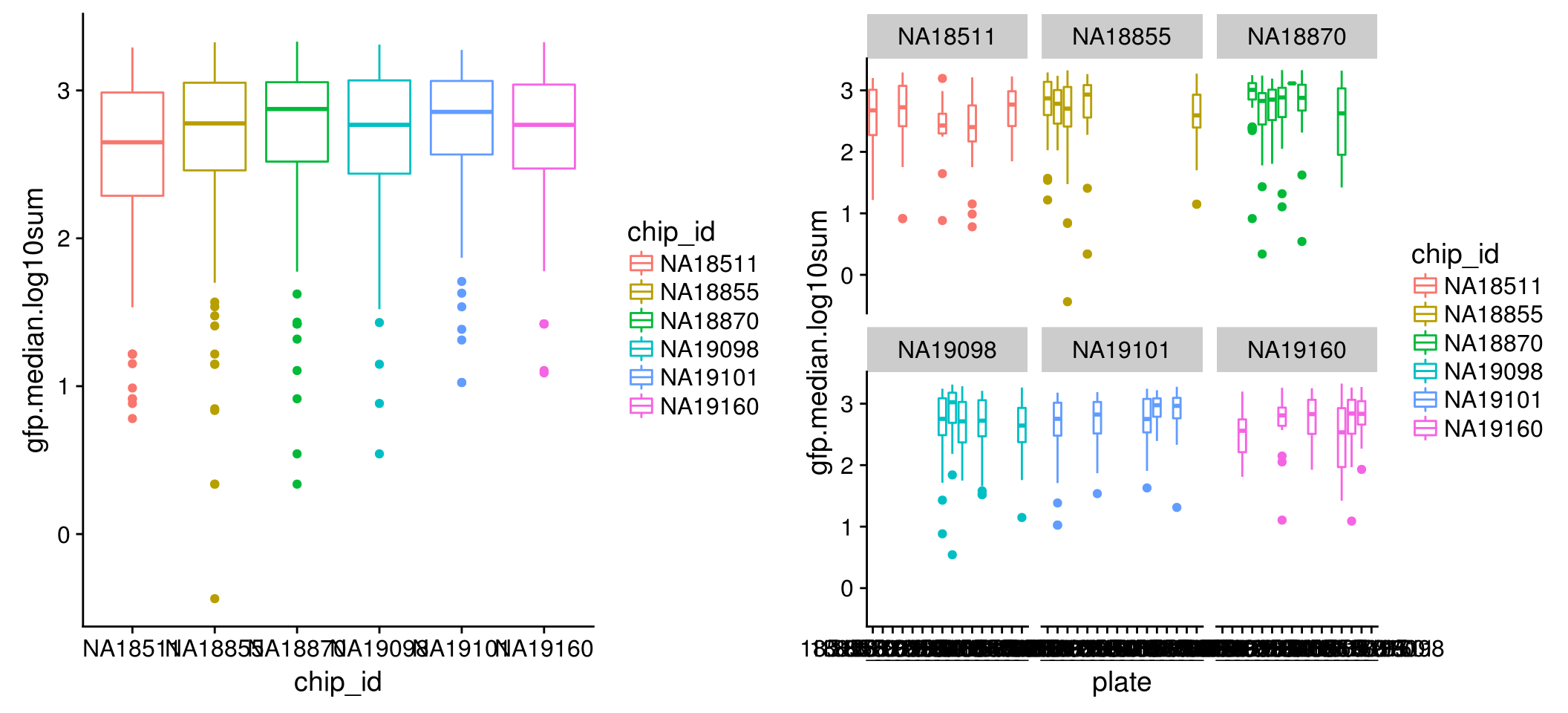
\(~\)
DAPI
\(~\)
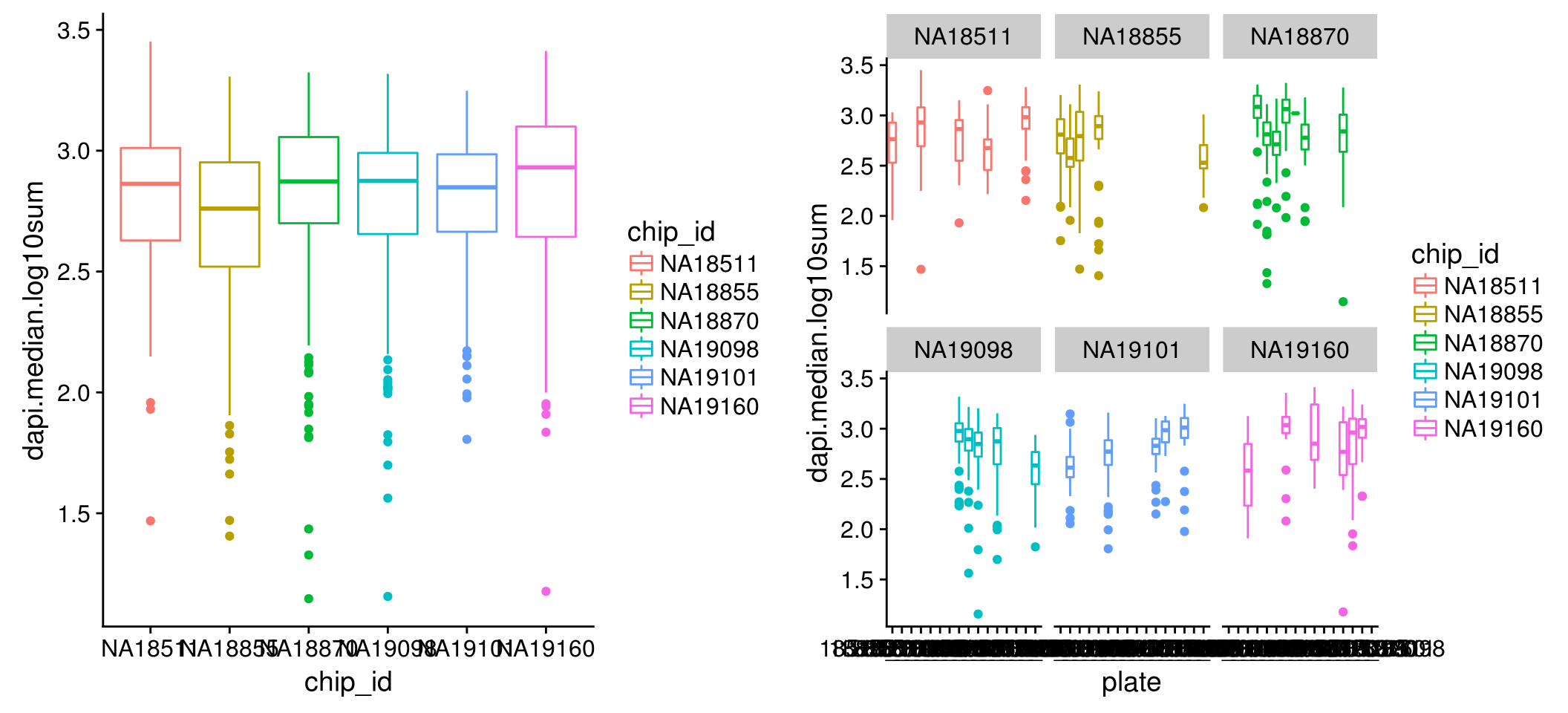
Intensity correlation
\(~\)
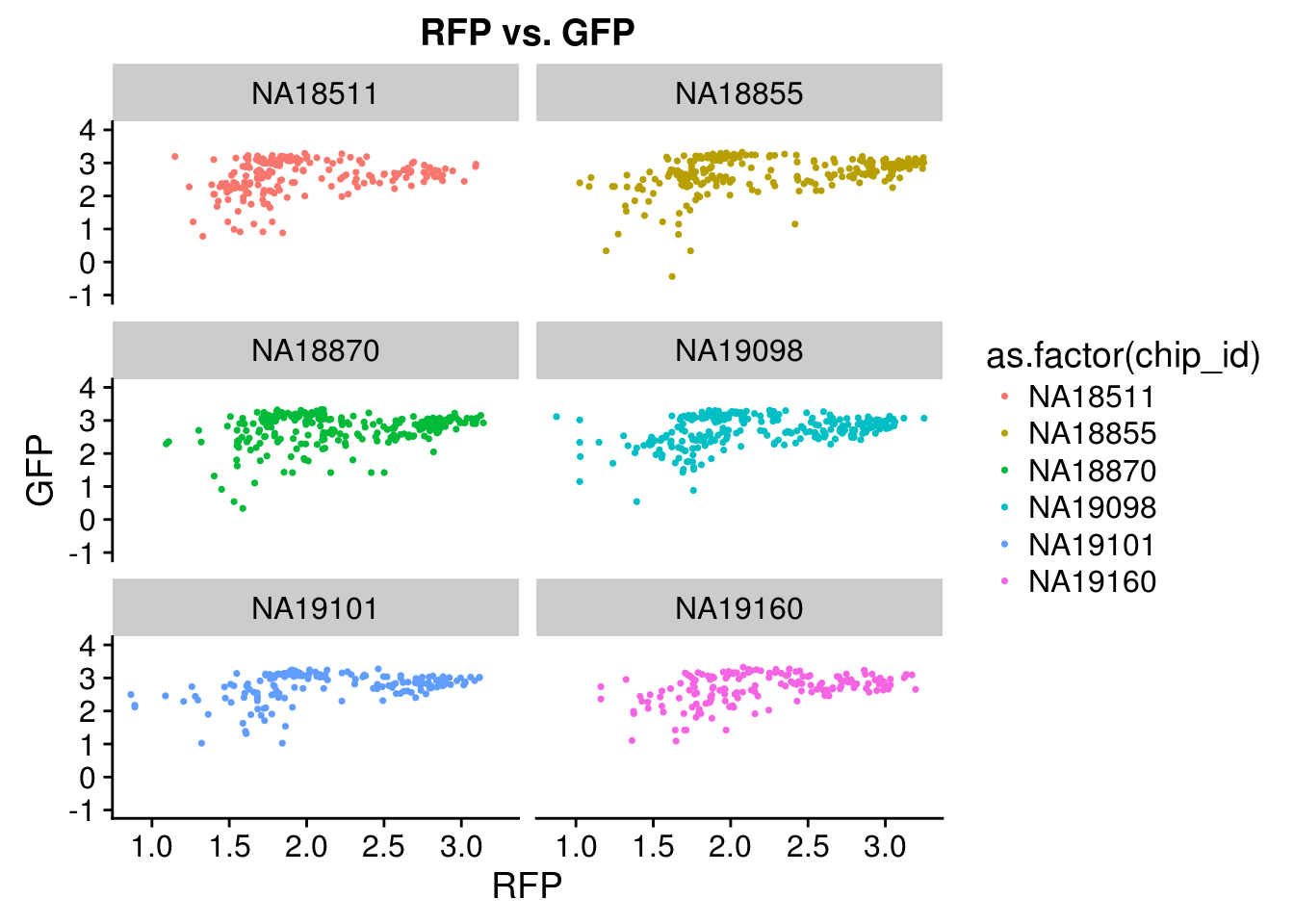
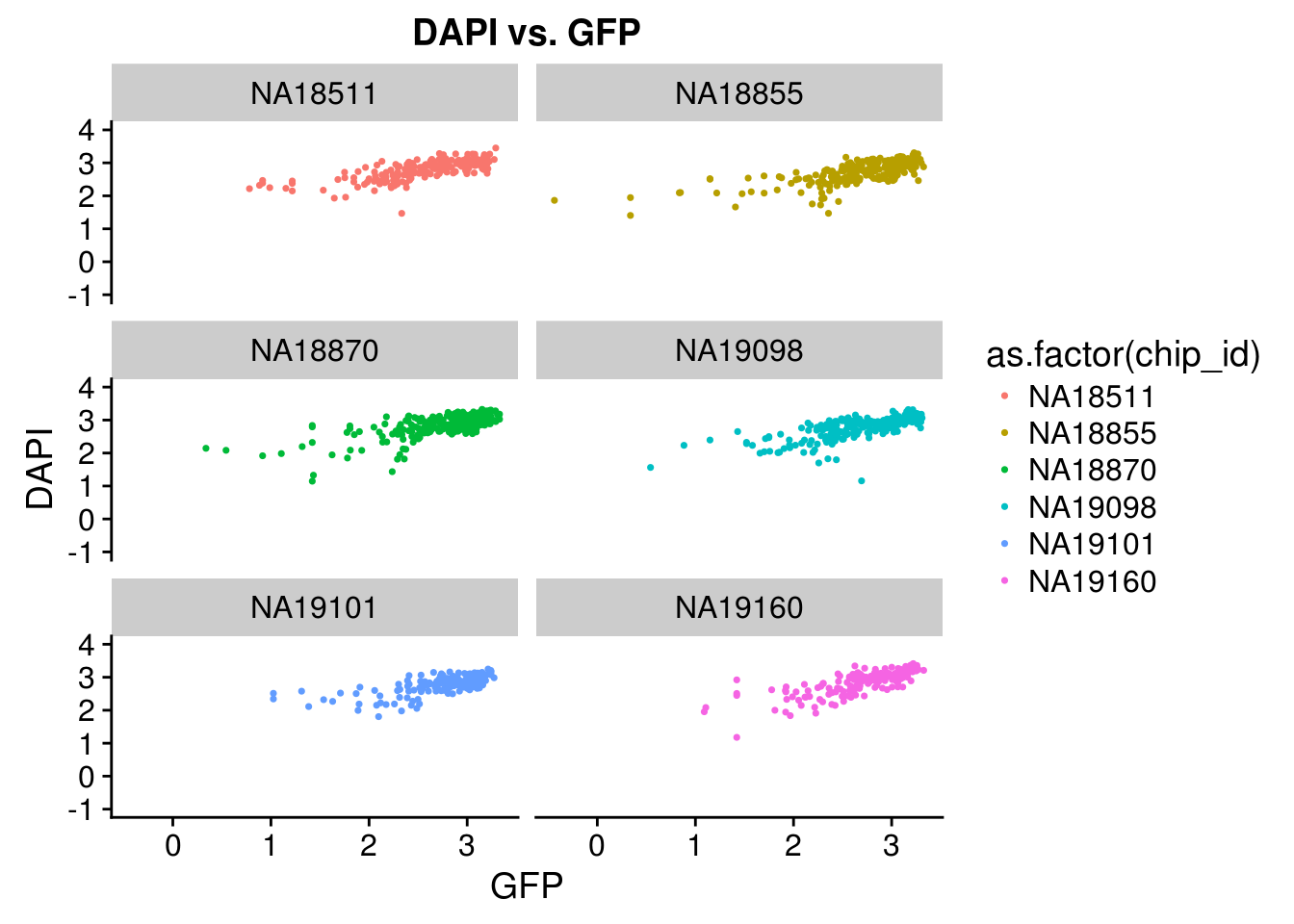
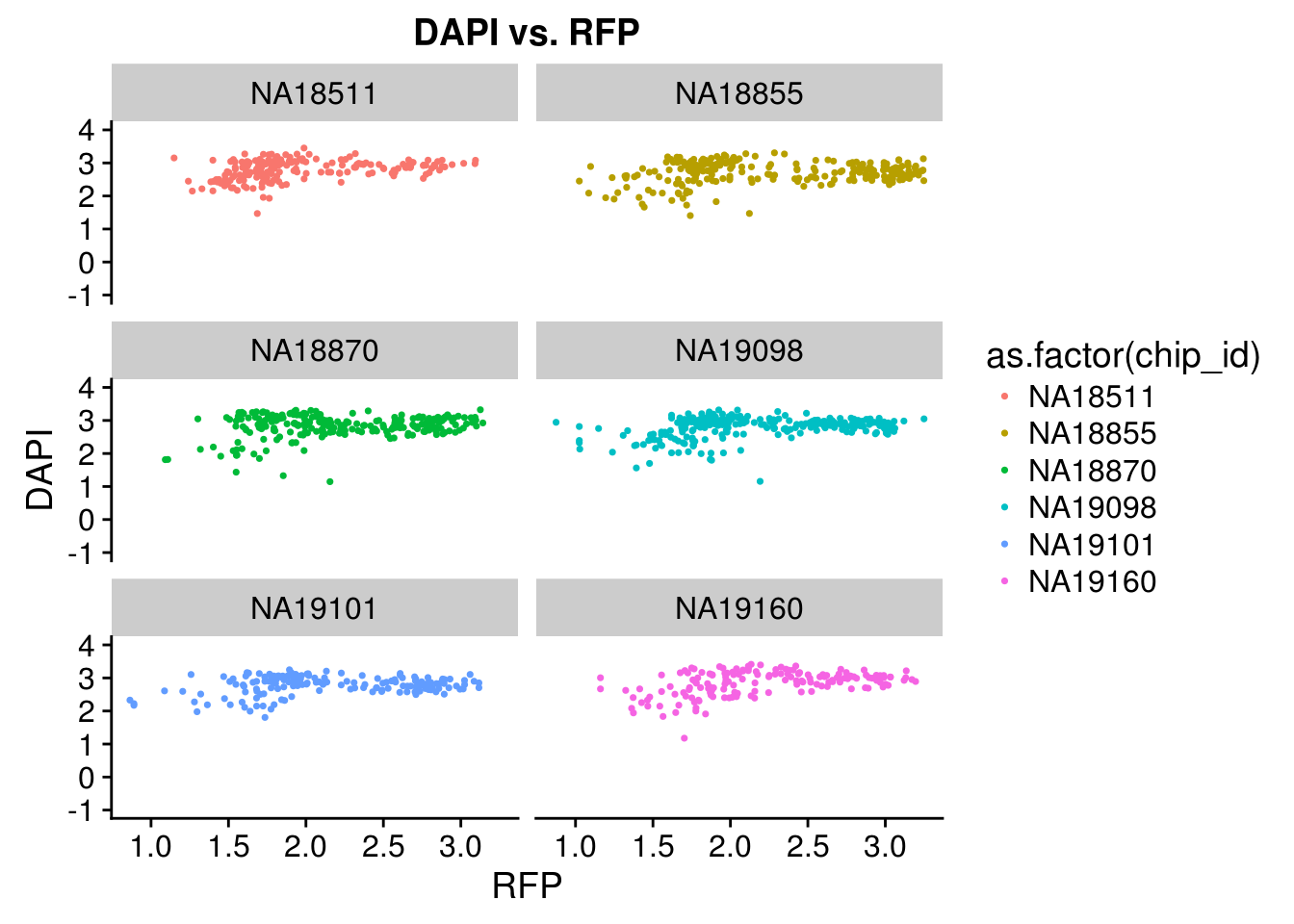
PCA
Proportion of variance explained by the 3 PCS.
[1] 0.63063931 0.27099047 0.09837022RFP versus PCs
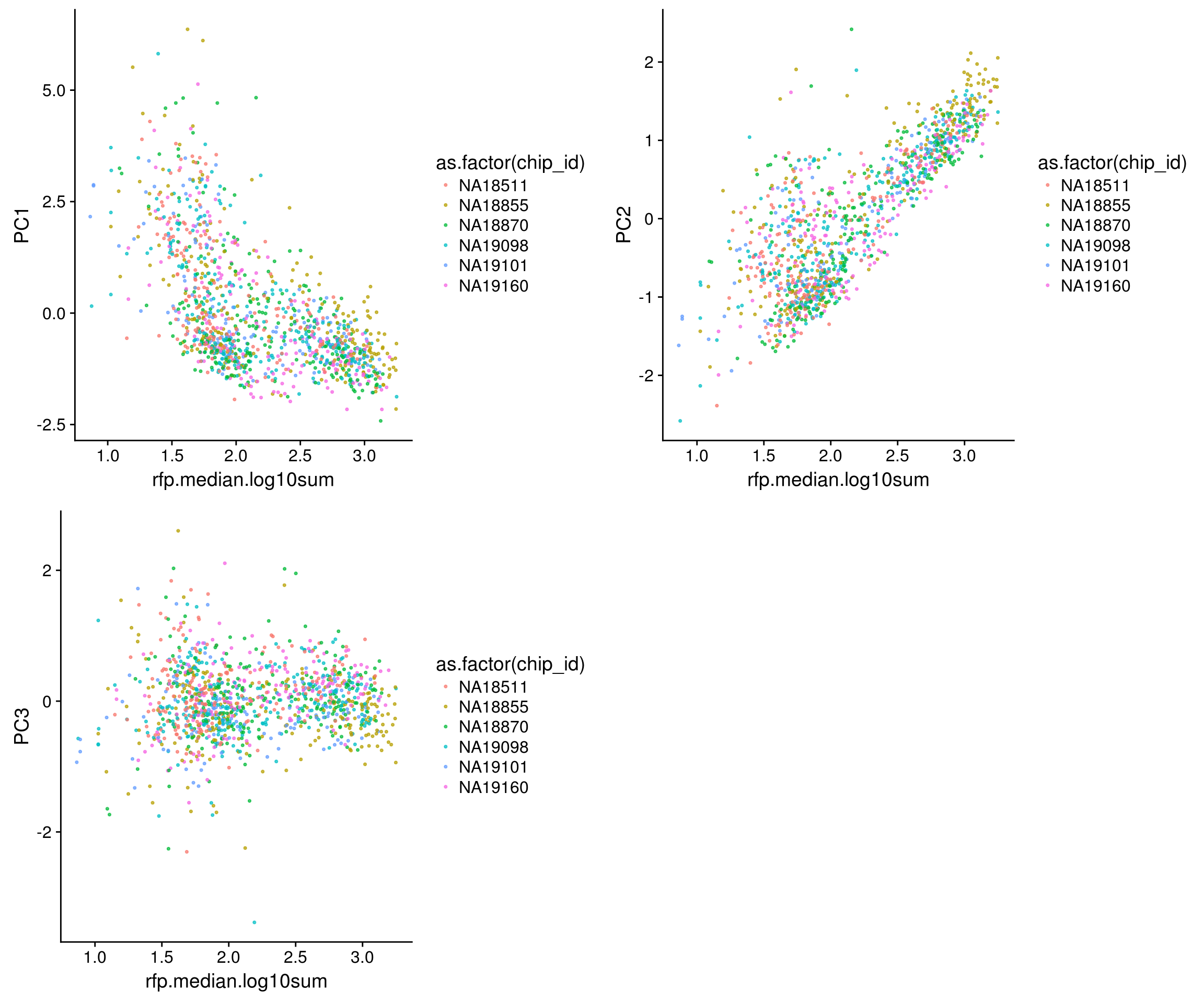
\(~\)
GFP versus PCs
\(~\)
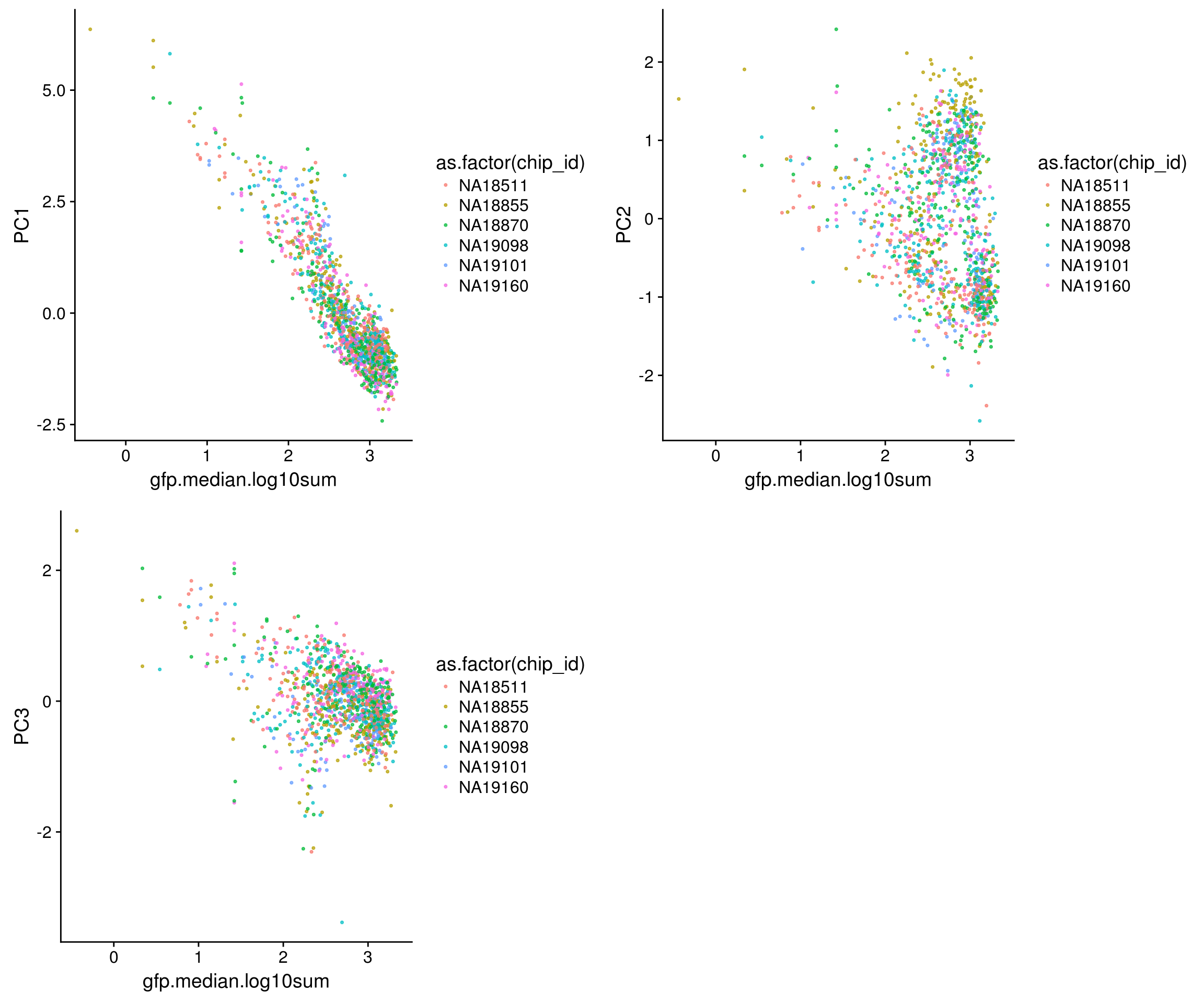
\(~\)
DAPI versus PCs
\(~\)
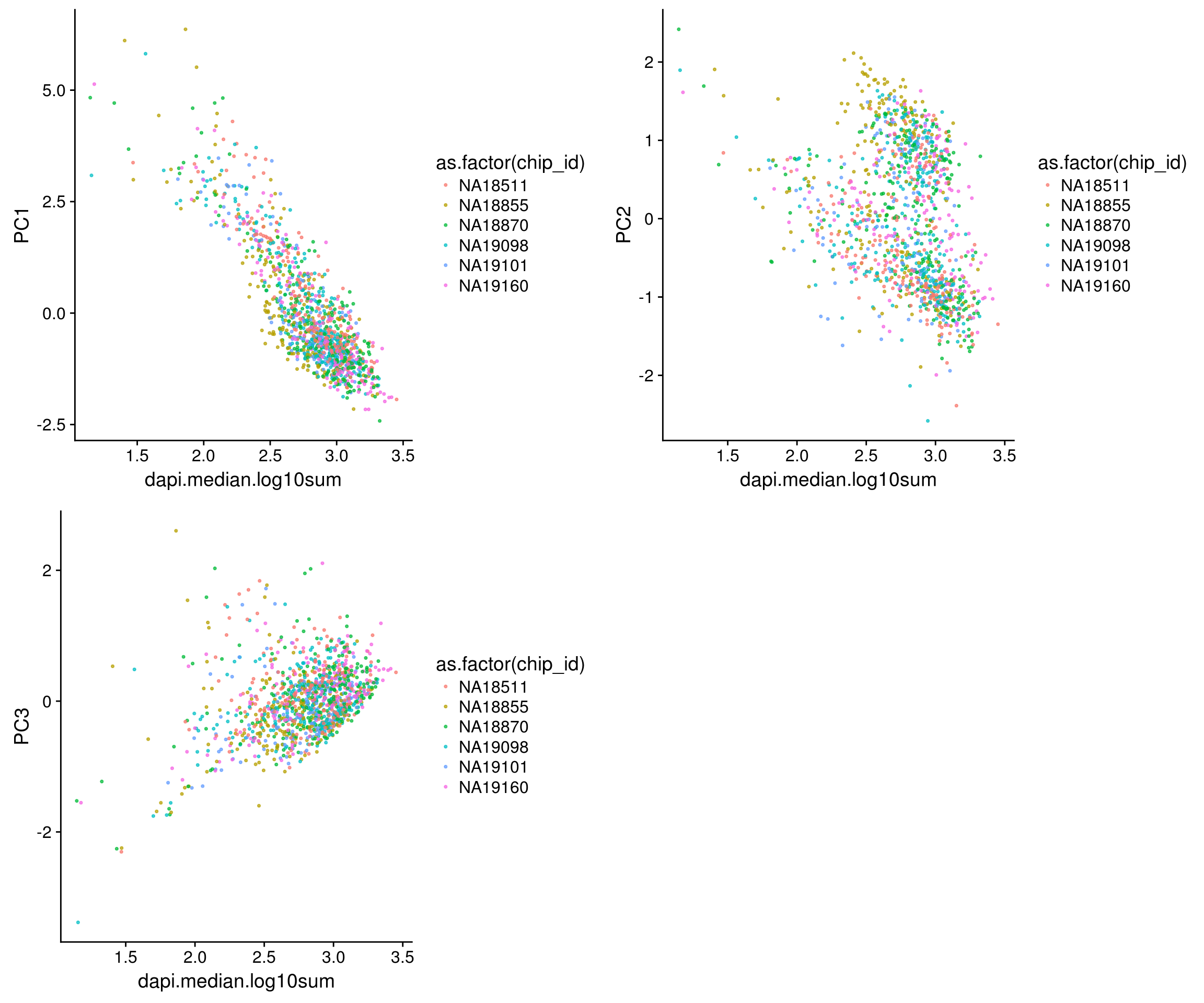
\(~\)
Plate versus PCs
\(~\)
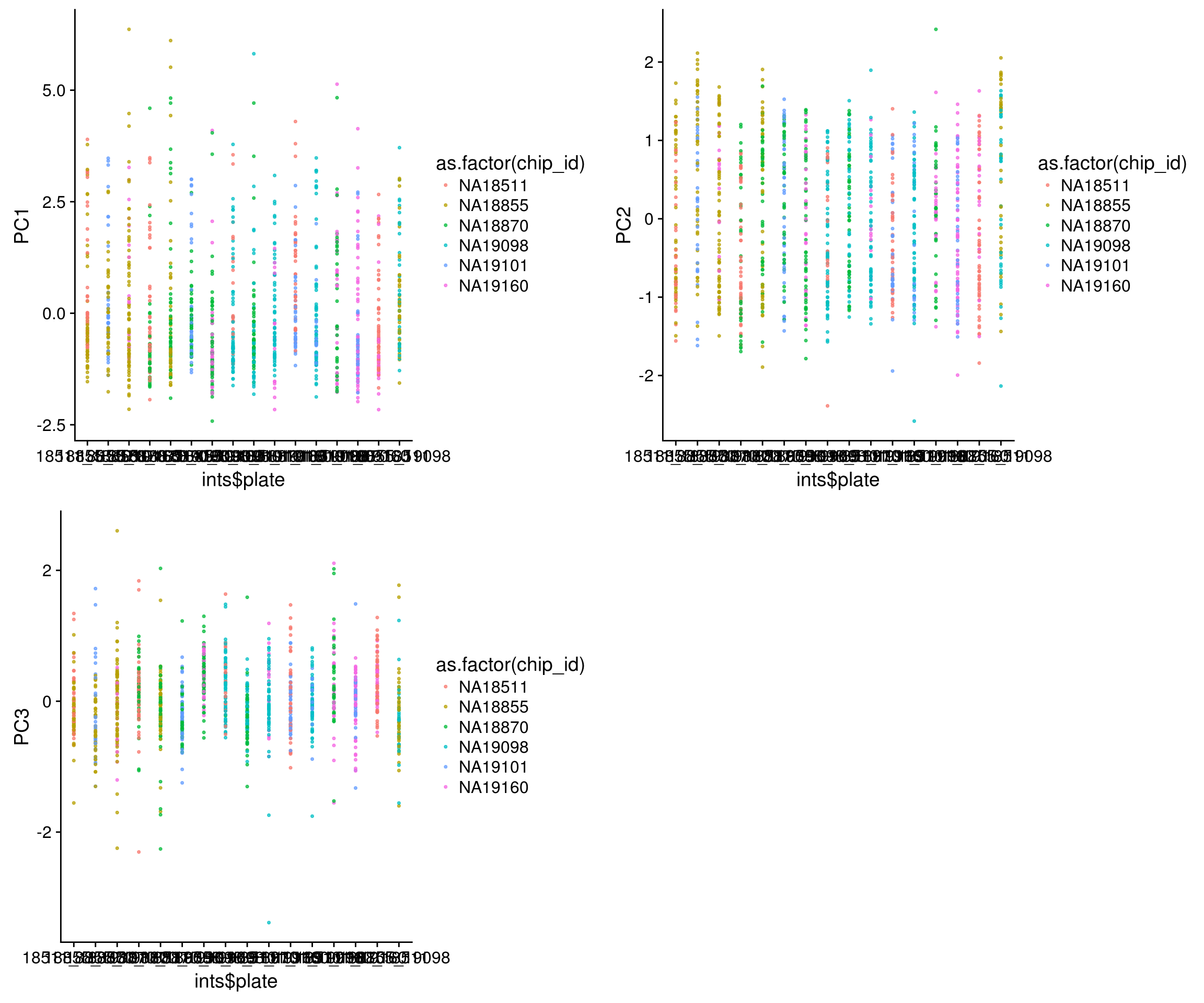
\(~\)
Individual label versus PCs
\(~\)
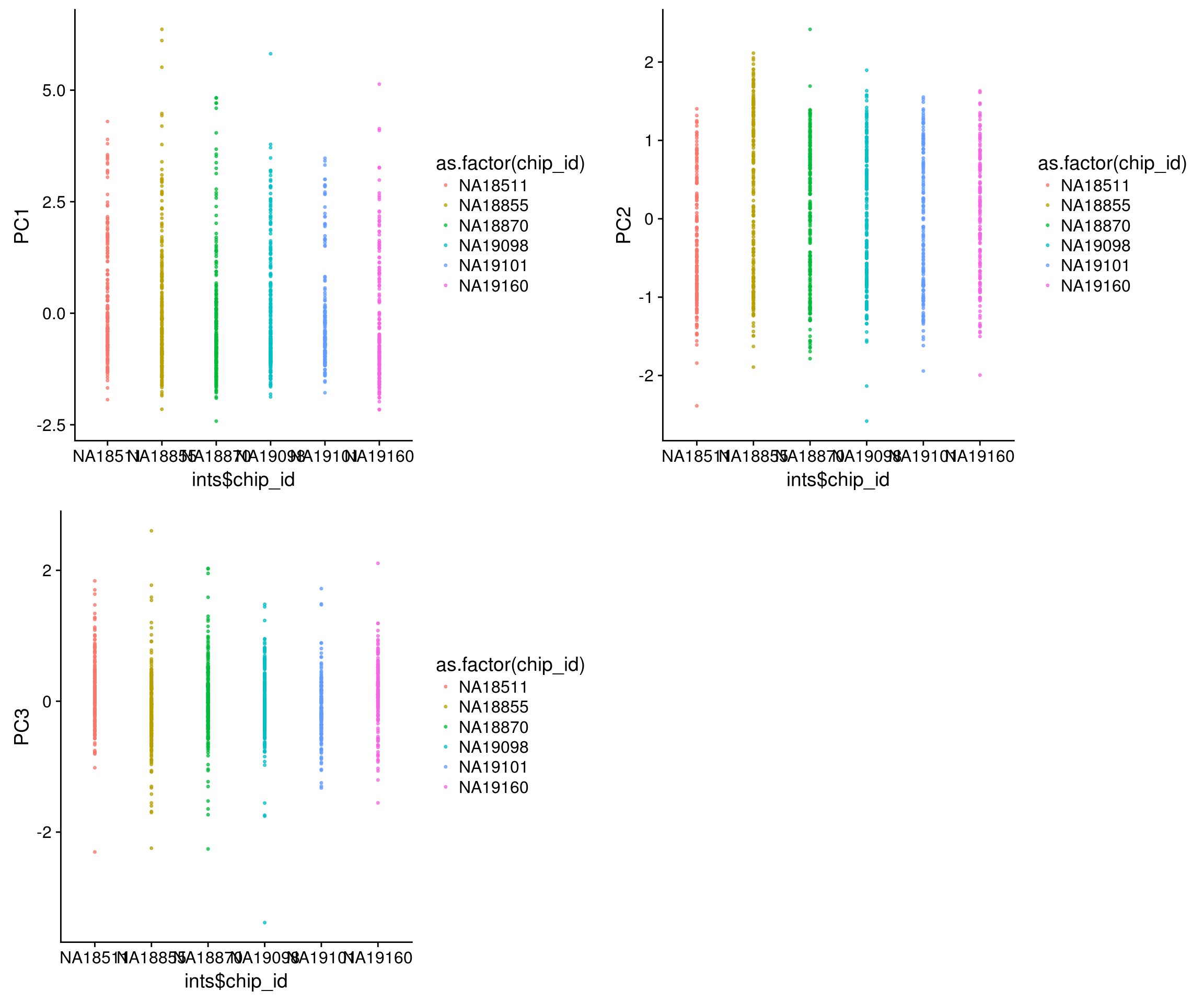
\(~\)
Nucleus size versus PCs
\(~\)
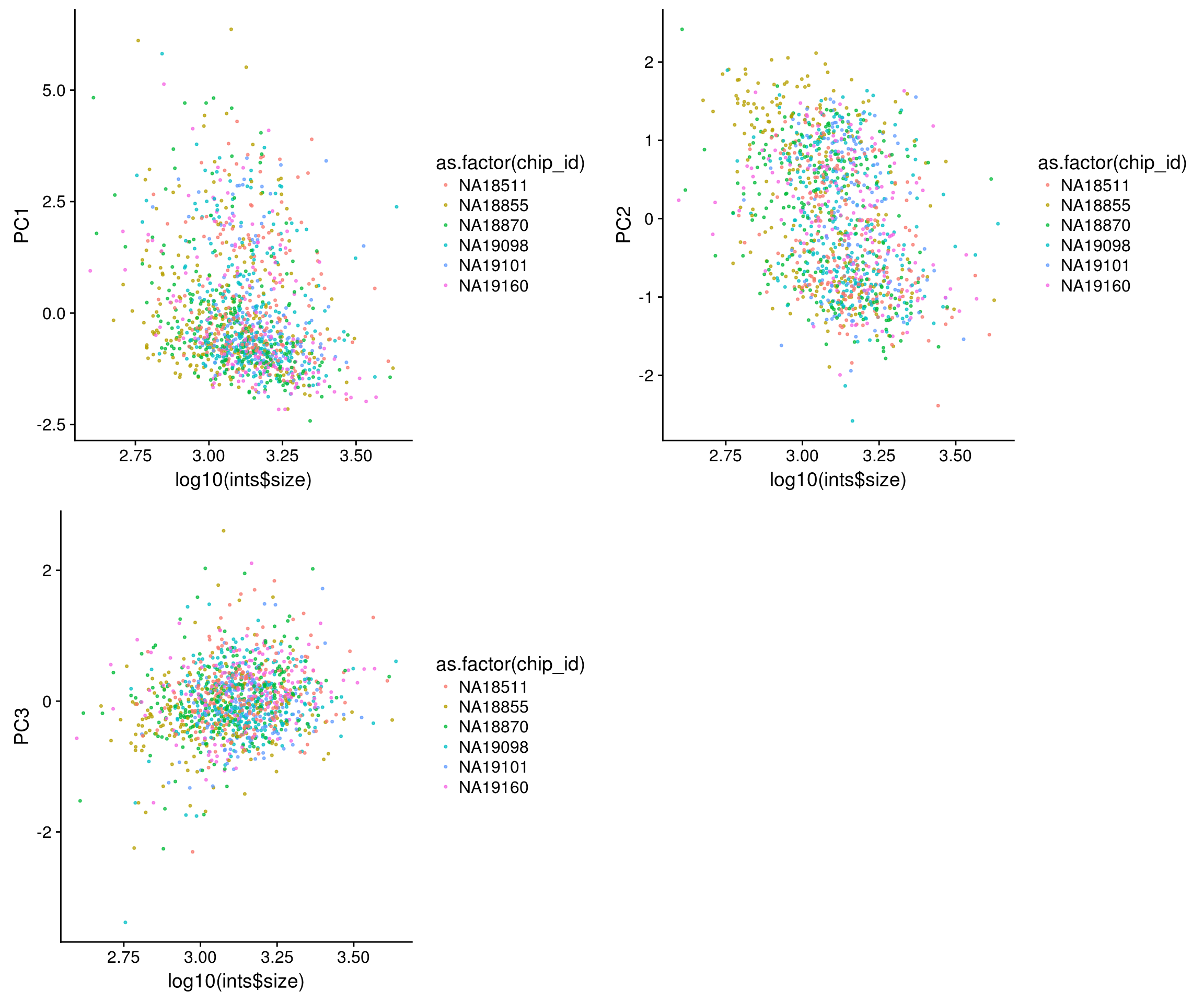
\(~\)
Eccentricity versus PCs
\(~\)
\(~\)
Perimeter versus PCs
\(~\)
\(~\)
Session information
R version 3.4.1 (2017-06-30)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.2 (Nitrogen)
Matrix products: default
BLAS: /home/joycehsiao/miniconda3/envs/fucci-seq/lib/R/lib/libRblas.so
LAPACK: /home/joycehsiao/miniconda3/envs/fucci-seq/lib/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] parallel stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] scales_0.4.1 Biobase_2.38.0 BiocGenerics_0.24.0
[4] RColorBrewer_1.1-2 wesanderson_0.3.2 cowplot_0.8.0
[7] ggplot2_2.2.1 dplyr_0.7.0 data.table_1.10.4
loaded via a namespace (and not attached):
[1] Rcpp_0.12.14 knitr_1.16 magrittr_1.5 munsell_0.4.3
[5] colorspace_1.3-2 R6_2.2.0 rlang_0.1.2 stringr_1.2.0
[9] plyr_1.8.4 tools_3.4.1 grid_3.4.1 gtable_0.2.0
[13] git2r_0.19.0 htmltools_0.3.6 lazyeval_0.2.0 yaml_2.1.14
[17] rprojroot_1.2 digest_0.6.12 assertthat_0.1 tibble_1.3.3
[21] glue_1.1.1 evaluate_0.10.1 rmarkdown_1.6 labeling_0.3
[25] stringi_1.1.2 compiler_3.4.1 backports_1.0.5 This R Markdown site was created with workflowr