Setting up the analysis folder
Create analysis
rrtools::use_analysis()✔ Adding bookdown to Imports
✔ Creating 'analysis' directory and contents
✔ Creating 'analysis'
✔ Creating 'analysis/paper'
✔ Creating 'analysis/figures'
✔ Creating 'analysis/templates'
✔ Creating 'analysis/data'
✔ Creating 'analysis/data/raw_data'
✔ Creating 'analysis/data/derived_data'
✔ Creating 'references.bib' from template.
✔ Creating 'paper.Rmd' from template.
Next, you need to: ↓ ↓ ↓ ↓
● Write your article/report/thesis, start at the paper.Rmd file
● Add the citation style library file (csl) to replace the default provided here, see https://github.com/citation-style-language/
● Add bibliographic details of cited items to the 'references.bib' file
● For adding captions & cross-referencing in an Rmd, see https://bookdown.org/yihui/bookdown/
● For adding citations & reference lists in an Rmd, see http://rmarkdown.rstudio.com/authoring_bibliographies_and_citations.html
Note that:
⚠ Your data files are tracked by Git and will be pushed to GitHubThis adds the following files:
├── analysis
│ ├── data
│ │ ├── DO-NOT-EDIT-ANY-FILES-IN-HERE-BY-HAND
│ │ ├── derived_data
│ │ └── raw_data
│ ├── figures
│ ├── paper
│ │ ├── paper.Rmd
│ │ └── references.bib
│ └── templates
│ ├── journal-of-archaeological-science.csl
│ ├── template.Rmd
│ └── template.docx
├── man
└── rrcompendium.RprojLet’s inspect
paper.Rmd
paper.Rmd is ready to write in and render with bookdown. It includes:
- a YAML header that identifies the
references.bibfile and the suppliedcslfile (to style the reference list) - a colophon that adds some git commit details to the end of the document. This means that the output file (HTML/PDF/Word) is always traceable to a specific state of the code.
references.bib
The references.bib file has just one item to demonstrate the format. It is ready to insert more reference details.
We can replace the supplied csl file with a different citation style from https://github.com/citation-style-language/
data/
Move gillespie.csv from the materials you downloaded in rrtools-wkshp-materials-master/ to analysis/data/raw_data/ in rrcompendium
Your data folder should now look like this:
analysis/data
├── DO-NOT-EDIT-ANY-FILES-IN-HERE-BY-HAND
├── derived_data
└── raw_data
└── gillespie.csvcreate journal article template using rticles
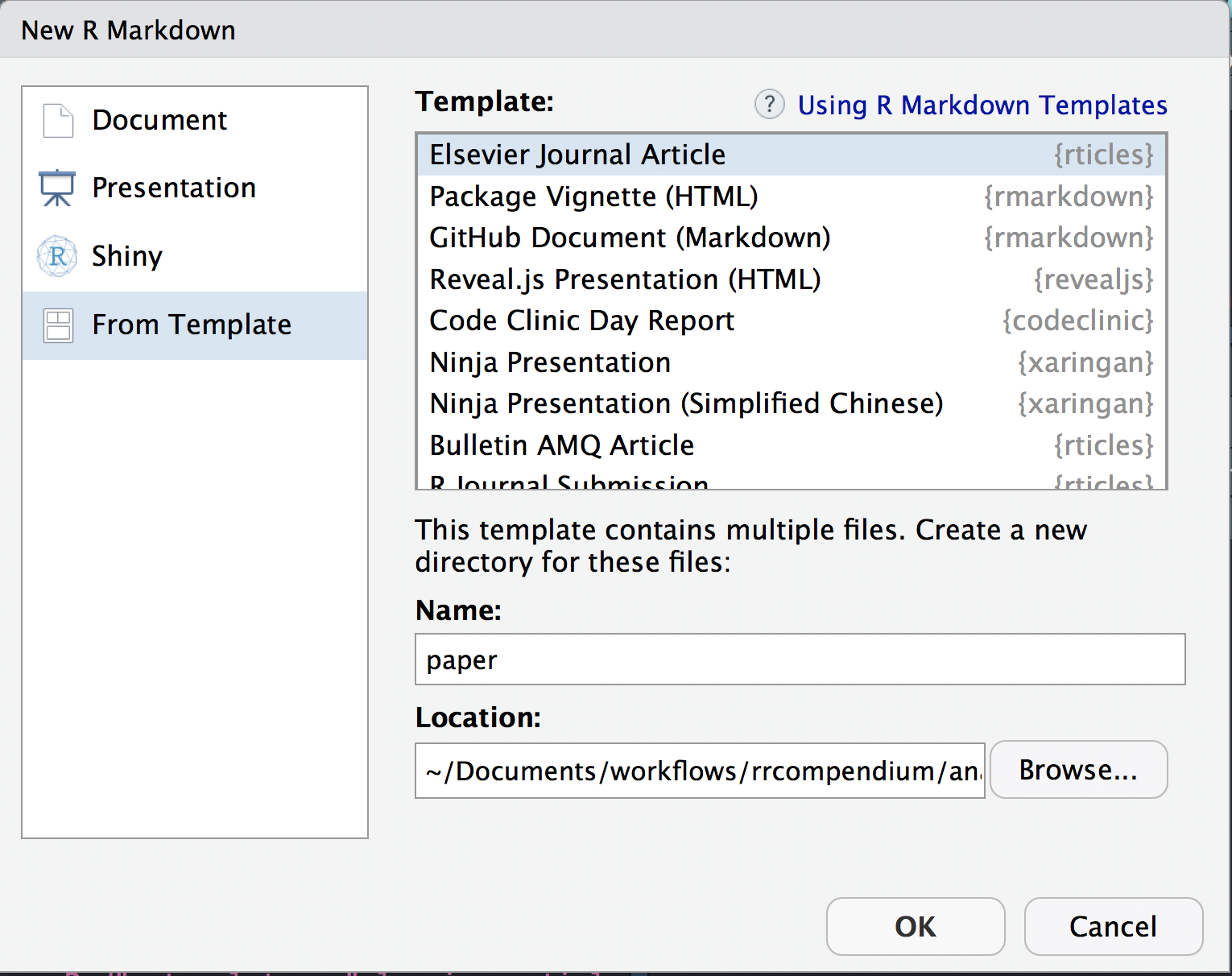
- Use the New R Markdown dialog to create an article from one of the templates. Select:
- Template: Elesevier Journal Article
- Name: paper
- Location:
~/Documents/workflows/rrcompendium/analysis
- Use the
rmarkdown::draft()function to create articles:
rmarkdown::draft(here::here("analysis","paper.Rmd"), template = "elsevier_article", package = "rticles")analysis/paper
├── elsarticle.cls
├── mybibfile.bib
├── numcompress.sty
└── paper.Rmd