Re-Analyze Efron’s Leukemia Data
Lei Sun
2018-04-16
Last updated: 2018-05-05
Code version: fbfd54d
source("../code/gdash_lik.R")
source("../code/gdfit.R")
library(limma)
library(locfdr)leukemia <- read.csv("http://web.stanford.edu/~hastie/CASI_files/DATA/leukemia_big.csv")
design <- c(rep(0, 20), rep(1, 14), rep(0, 27), rep(1, 11))
lim = limma::lmFit(leukemia, model.matrix(~design))
r.ebayes = limma::eBayes(lim)
p = r.ebayes$p.value[, 2]
t = r.ebayes$t[, 2]
z = -sign(t) * qnorm(p/2)
betahat = lim$coefficients[,2]
sebetahat = betahat / zhist(p)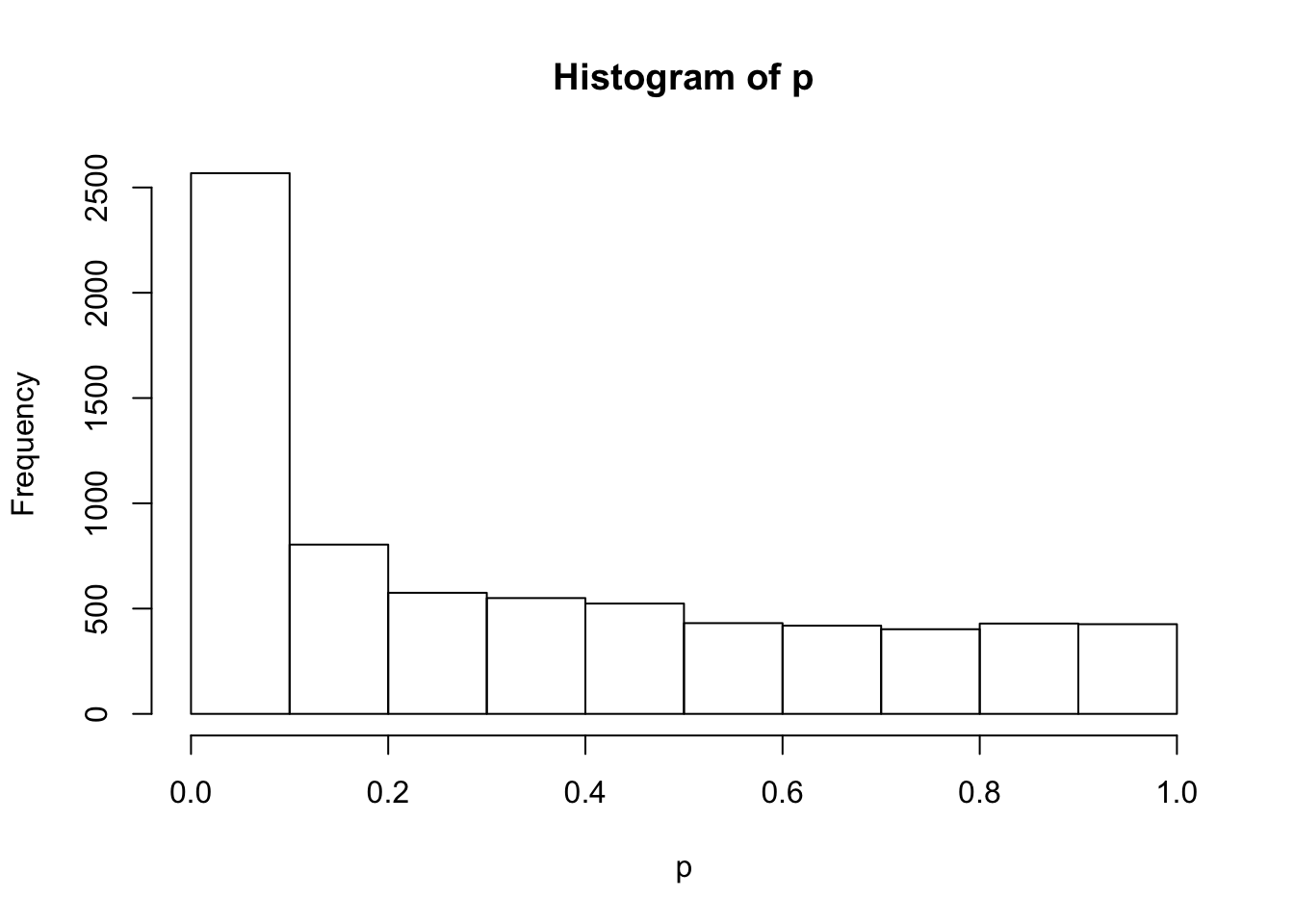
hist(z, prob = TRUE, ylim = c(0, dnorm(0)))
lines(seq(-10, 10, by = 0.01), dnorm(seq(-10, 10, by = 0.01)), col = "red")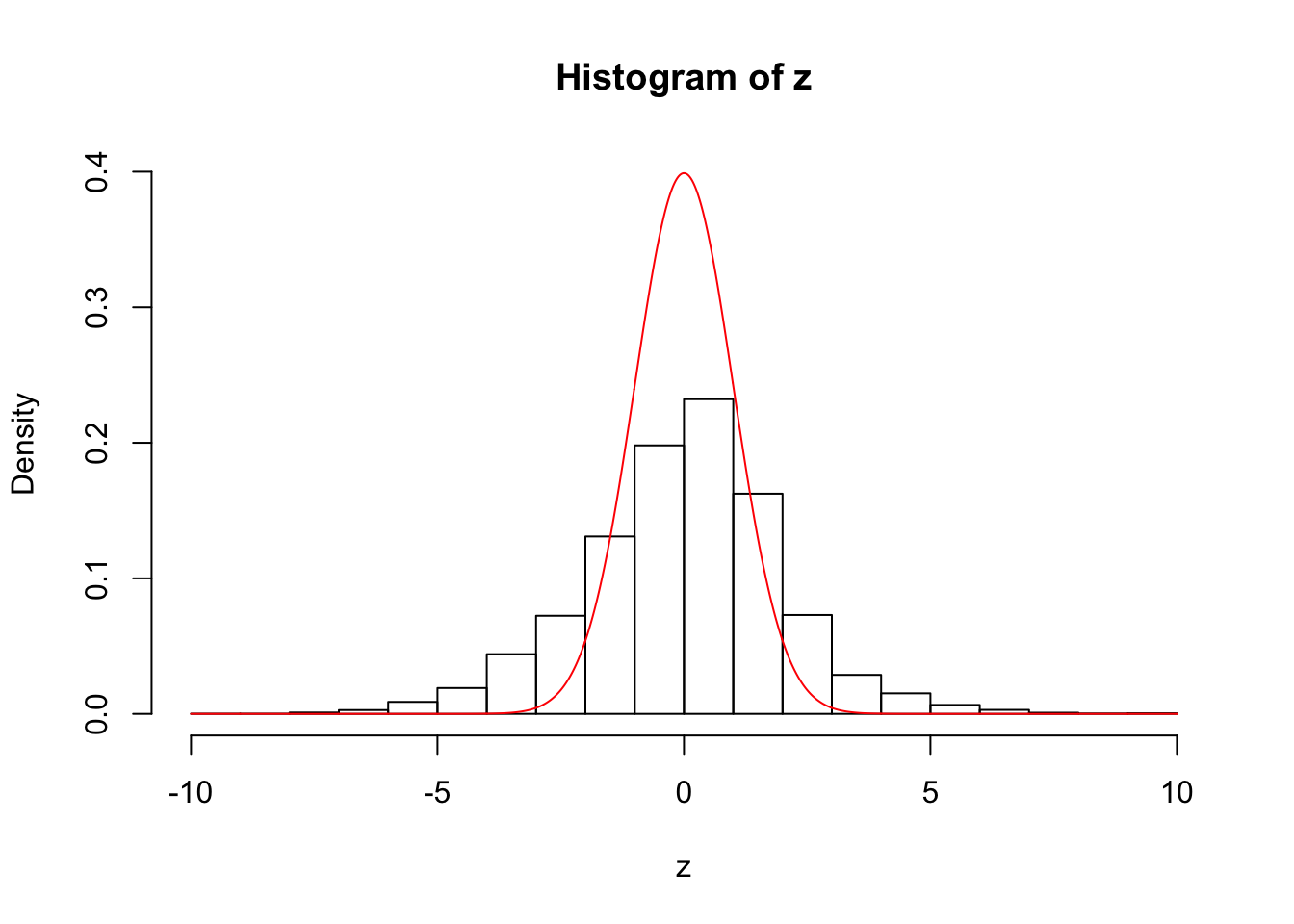
fit <- gdash(betahat, sebetahat, gd.ord = 10)
# rand.sign <- sample(c(rep(1, nrow(leukemia) / 2), rep(-1, nrow(leukemia) / 2)))
# fit.sym <- gdash(rand.sign * betahat, sebetahat, gd.ord = 10)
fit.locfdr <- locfdr(z)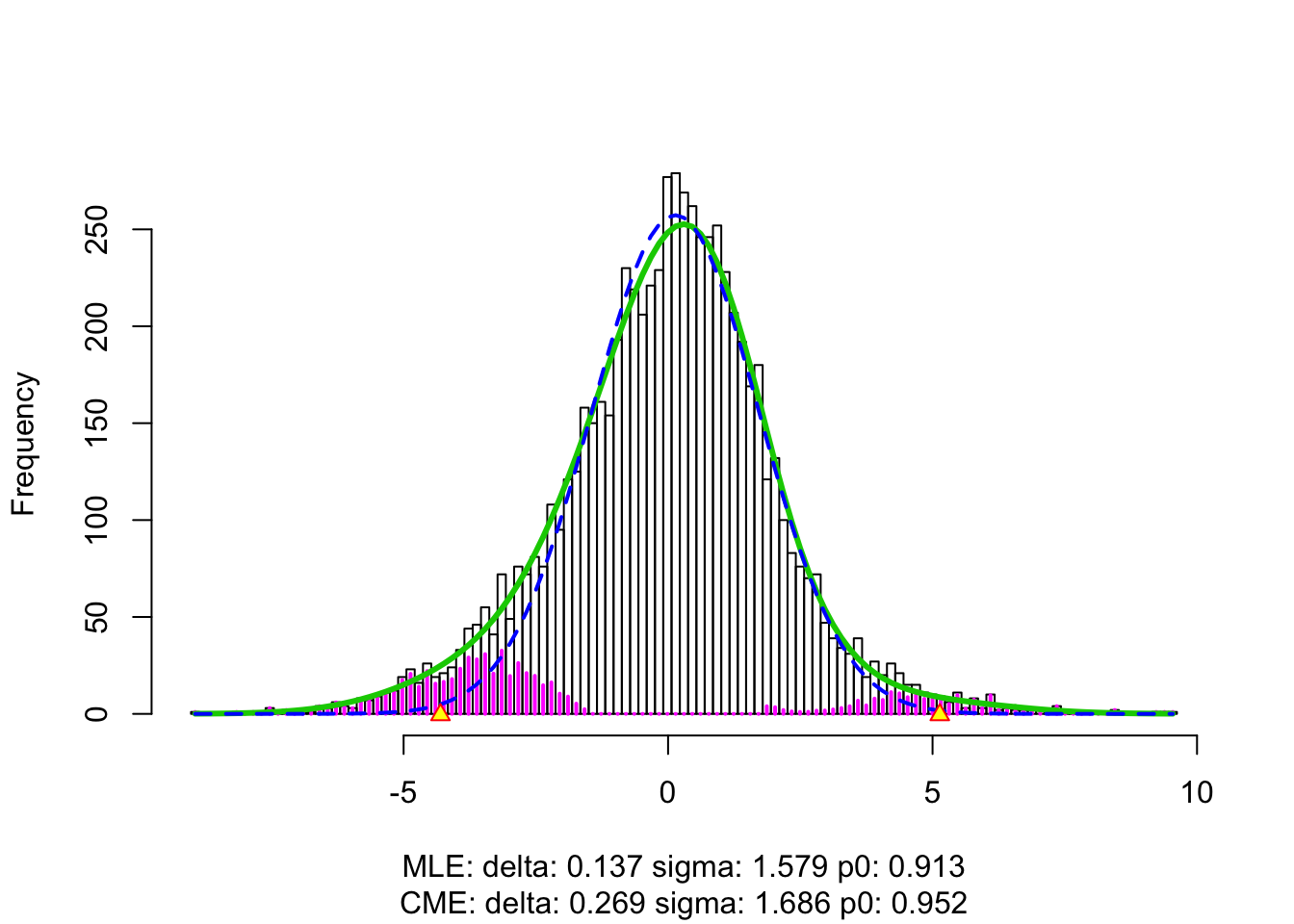
fit.ash <- ashr::ash(betahat, sebetahat, mixcompdist = "normal", method = "fdr")
fit.qvalue <- qvalue::qvalue(p)
hist(z[abs(z) >= 5])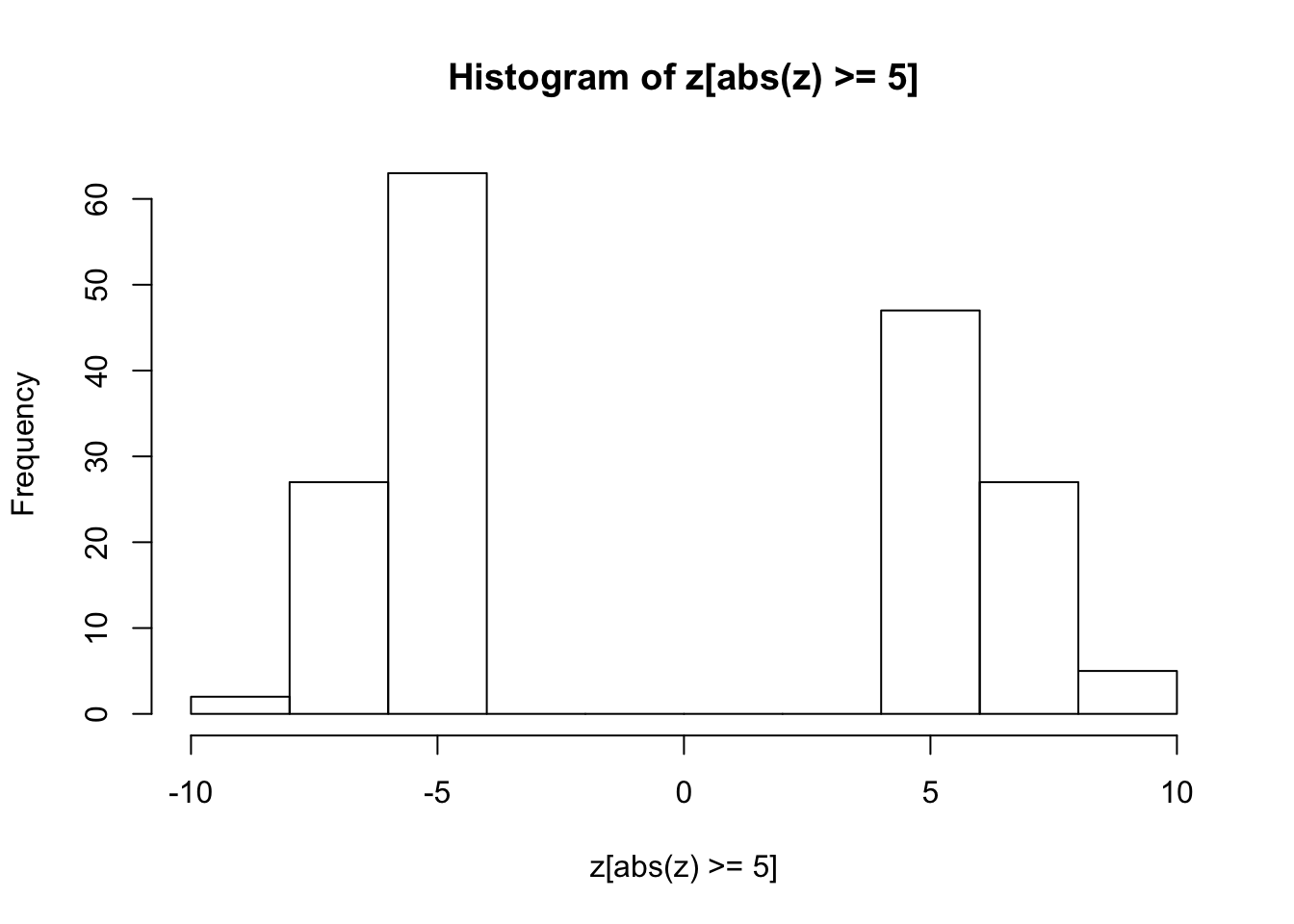
hist(betahat[abs(z) >= 5])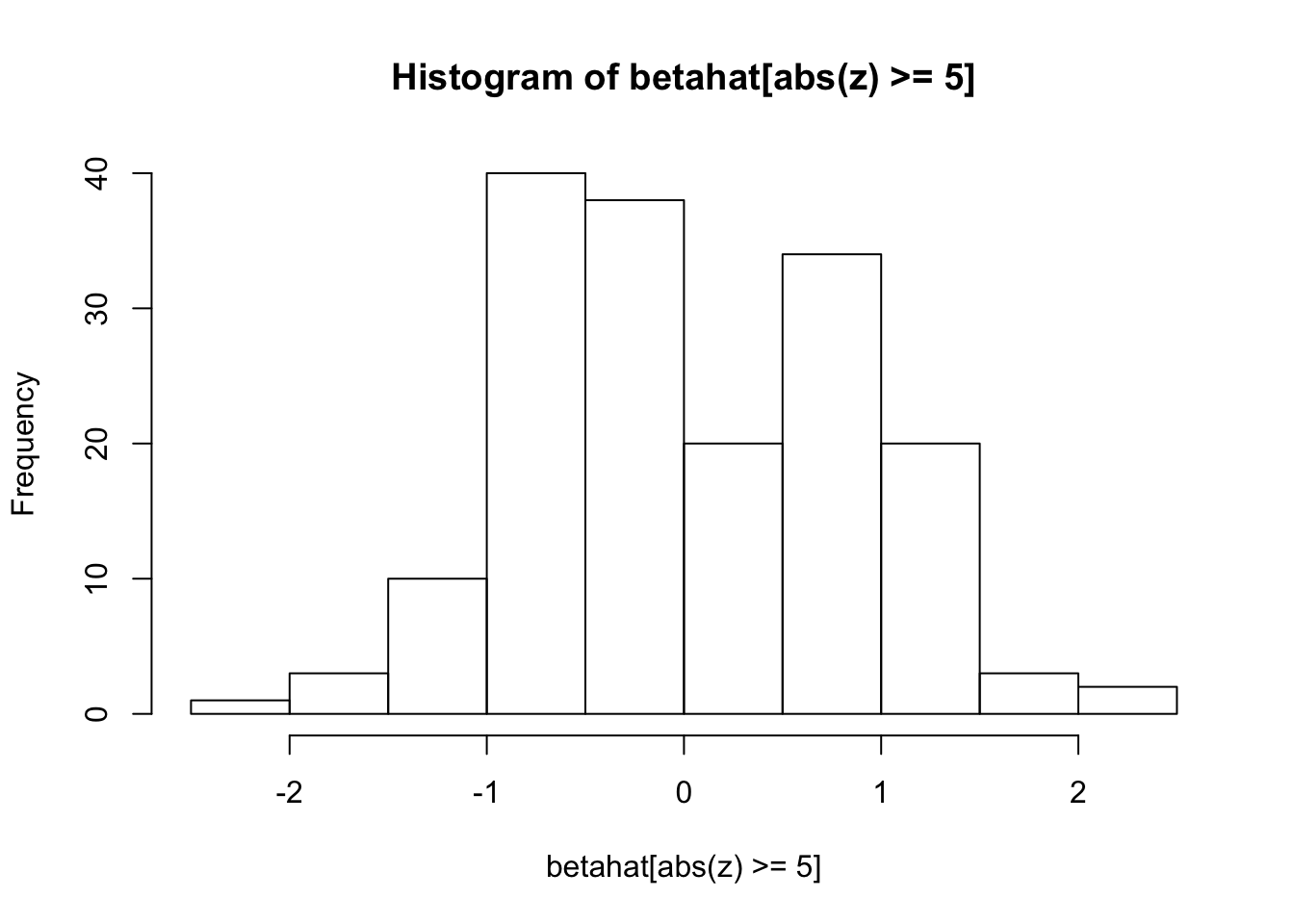
x.plot <- seq(-2, 2, length = 1000)
# y.plot <- sapply(fit.sym$fitted_g$sd, function(x) {sapply(x.plot, dnorm, sd = x)})
# y.plot <- y.plot %*% fit.sym$fitted_g$pi
# plot(x.plot, y.plot, type = "l", col = "red")
# y.plot <- sapply(fit$fitted_g$sd, function(x) {sapply(x.plot, dnorm, sd = x)})
# y.plot <- y.plot %*% fit$fitted_g$pi
# lines(x.plot, y.plot)
x.plot <- seq(-10, 10, length = 1000)
gd.ord <- 10
hermite = Hermite(gd.ord)
gd0.std = dnorm(x.plot)
matrix_lik_plot = cbind(gd0.std)
for (i in 1 : gd.ord) {
gd.std = (-1)^i * hermite[[i]](x.plot) * gd0.std / sqrt(factorial(i))
matrix_lik_plot = cbind(matrix_lik_plot, gd.std)
}
y.plot = matrix_lik_plot %*% fit$w * fit$fitted_g$pi[1]
library(scales)
method.col <- hue_pal()(5)
par(mfrow = c(1, 2))
hist(z, prob = TRUE, main = "", xlab = expression(paste(z, "-scores")))
lines(x.plot, y.plot, col = method.col[5], lwd = 2)
lines(x.plot, dnorm(x.plot), col =
"orange"
# method.col[2]
, lty = 2, lwd = 2)
lines(x.plot, dnorm(x.plot, fit.locfdr$fp0[3, 1], fit.locfdr$fp0[3, 2]) * fit.locfdr$fp0[3, 3], col = method.col[3], lty = 2, lwd = 2)
text(-3, 0.22, "N(0,1)", col = "orange")
text(-5, 0.15, bquote(atop("Efron's empirical null", .(round(fit.locfdr$fp0[3, 3], 2)) %*% N(.(round(fit.locfdr$fp0[3, 1], 2)), .(round(fit.locfdr$fp0[3, 2], 2))^2))), col = method.col[3])
text(4.5, 0.10, "CASH null", col = method.col[5])
arrows(-1.8, 0.22, -1.1, 0.215, length = 0.1, angle = 20, col = "orange")
arrows(-2, 0.15, -1.5, 0.145, length = 0.1, angle = 20, col = method.col[3])
arrows(2.9, 0.10, 2, 0.095, length = 0.1, angle = 20, col = method.col[5])
plot(z, fit$lfdr, pch = 18, cex = 0.5, ylim = c(0, 1), col = method.col[5], ylab = "Local FDR", xlab = expression(paste(z, "-scores")))
points(z, fit.locfdr$fdr, pch = 16, cex = 0.5, col = method.col[3])
points(z, ashr::get_lfdr(fit.ash), pch = 17, cex = 0.5, col = method.col[4])
points(z, fit.qvalue$lfdr, pch = 15, cex = 0.5, col = method.col[2])
abline(h = 0.2, lty = 2)
legend("topright", bty = "n", pch = 15 : 18, col = method.col[2 : 5], c("qvalue", "locfdr", "ASH", "CASH"))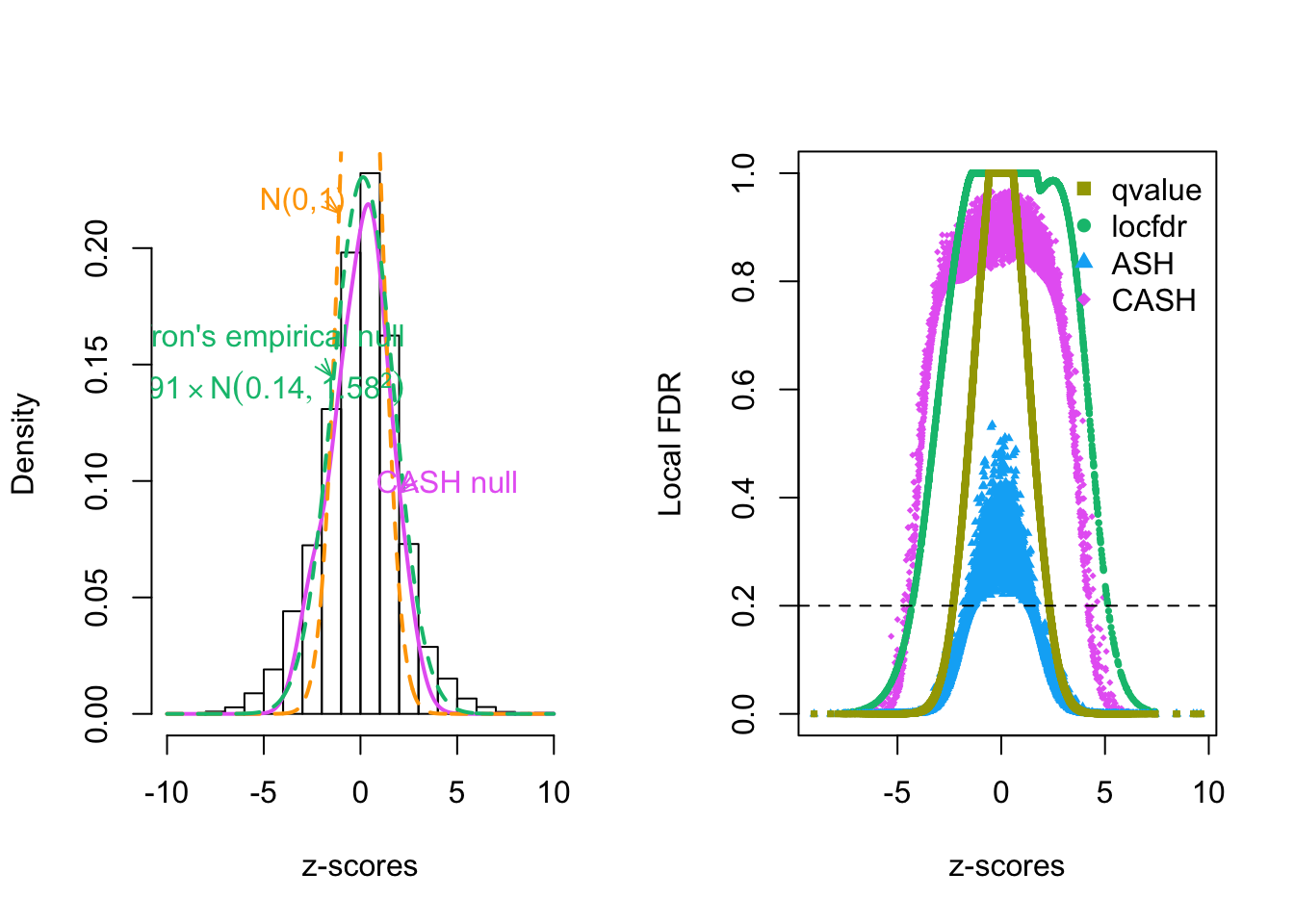
library(lattice)
test <- seq(z)[fit$lfdr <= 0.1 & fit.locfdr$fdr > 0.1]
hist(z, prob = TRUE)
hist(z[test], xlim = c(-10, 10))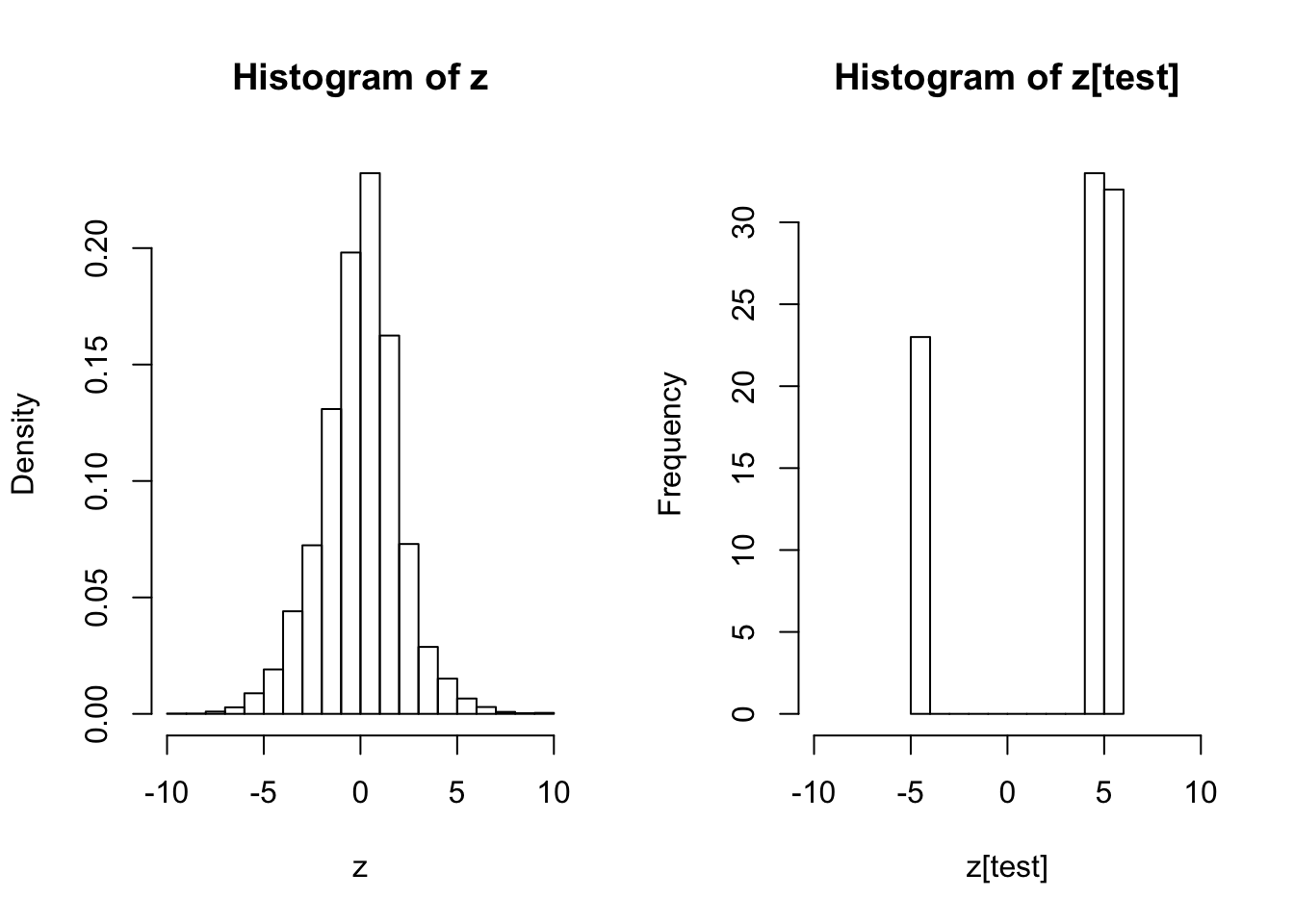
betahat <- c()
p <- c()
z <- c()
t <- c()
leukemia.rank <- apply(leukemia, 2, function(x) {qnorm((rank(x) - 0.5) / 7128)})
for (i in seq(nrow(leukemia))) {
ttest <- t.test(leukemia[i, ][design == 0], leukemia[i, ][design == 1], var.equal = TRUE)
betahat[i] <- ttest$estimate[1] - ttest$estimate[2]
t[i] <- ttest$statistic
p[i] <- ttest$p.value
z[i] <- -qnorm(ttest$p.value / 2) * sign(ttest$statistic)
}
z1 <- qnorm(pt(t, 70))
z2 <- -qnorm(pt(-t, 70))
z <- z1
z[abs(z1) > abs(z2)] <- z2[abs(z1) > abs(z2)]
sebetahat <- betahat / zprostate <- read.csv("http://web.stanford.edu/~hastie/CASI_files/DATA/prostmat.csv")
design <- c(rep(0, 50), rep(1, 52))
lim = limma::lmFit(prostate, model.matrix(~design))
r.ebayes = limma::eBayes(lim)
p = r.ebayes$p.value[, 2]
t = r.ebayes$t[, 2]
z = -sign(t) * qnorm(p/2)
betahat = lim$coefficients[,2]
sebetahat = betahat / zhist(p)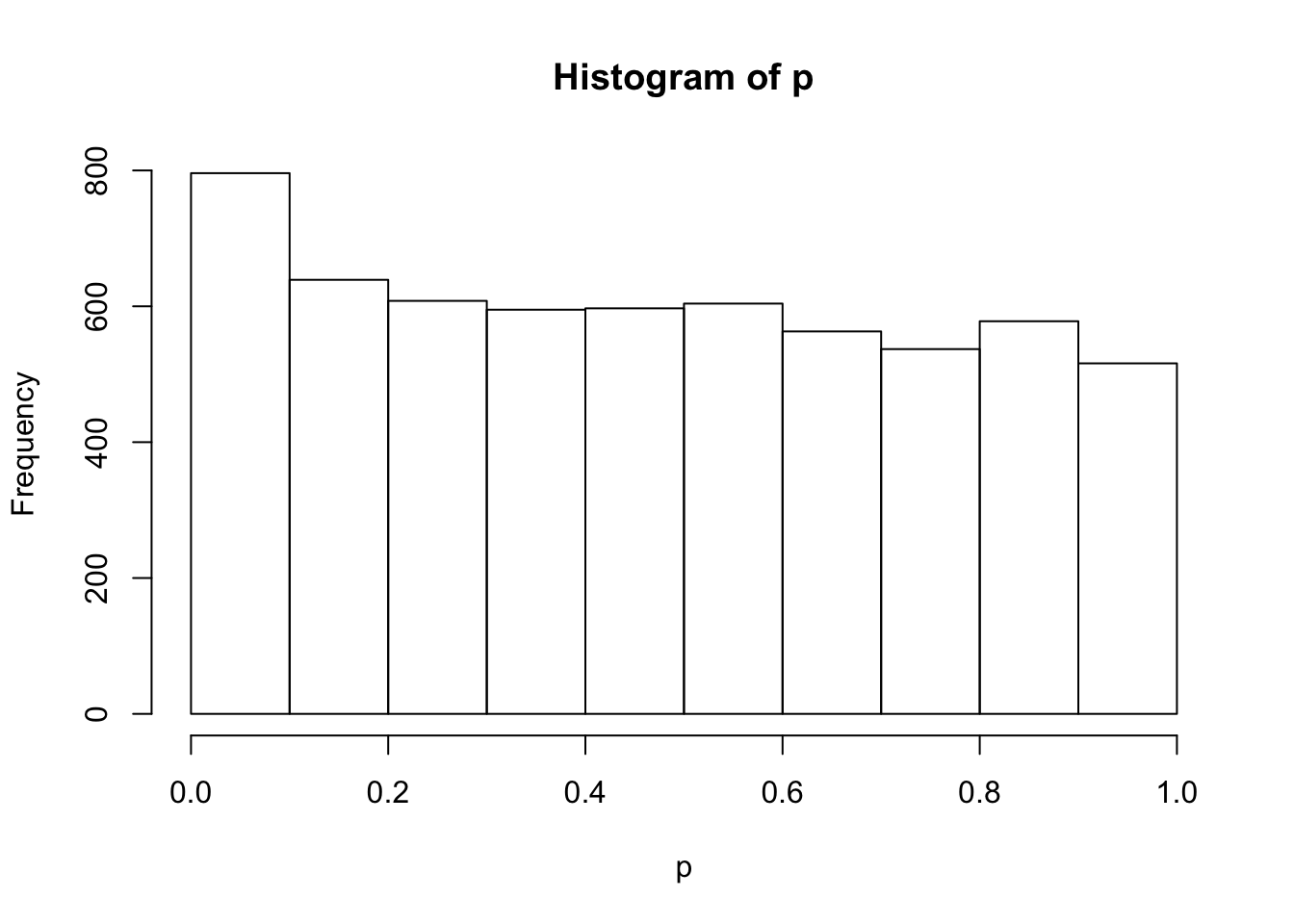
hist(z, prob = TRUE, ylim = c(0, dnorm(0)))
lines(seq(-10, 10, by = 0.01), dnorm(seq(-10, 10, by = 0.01)), col = "red")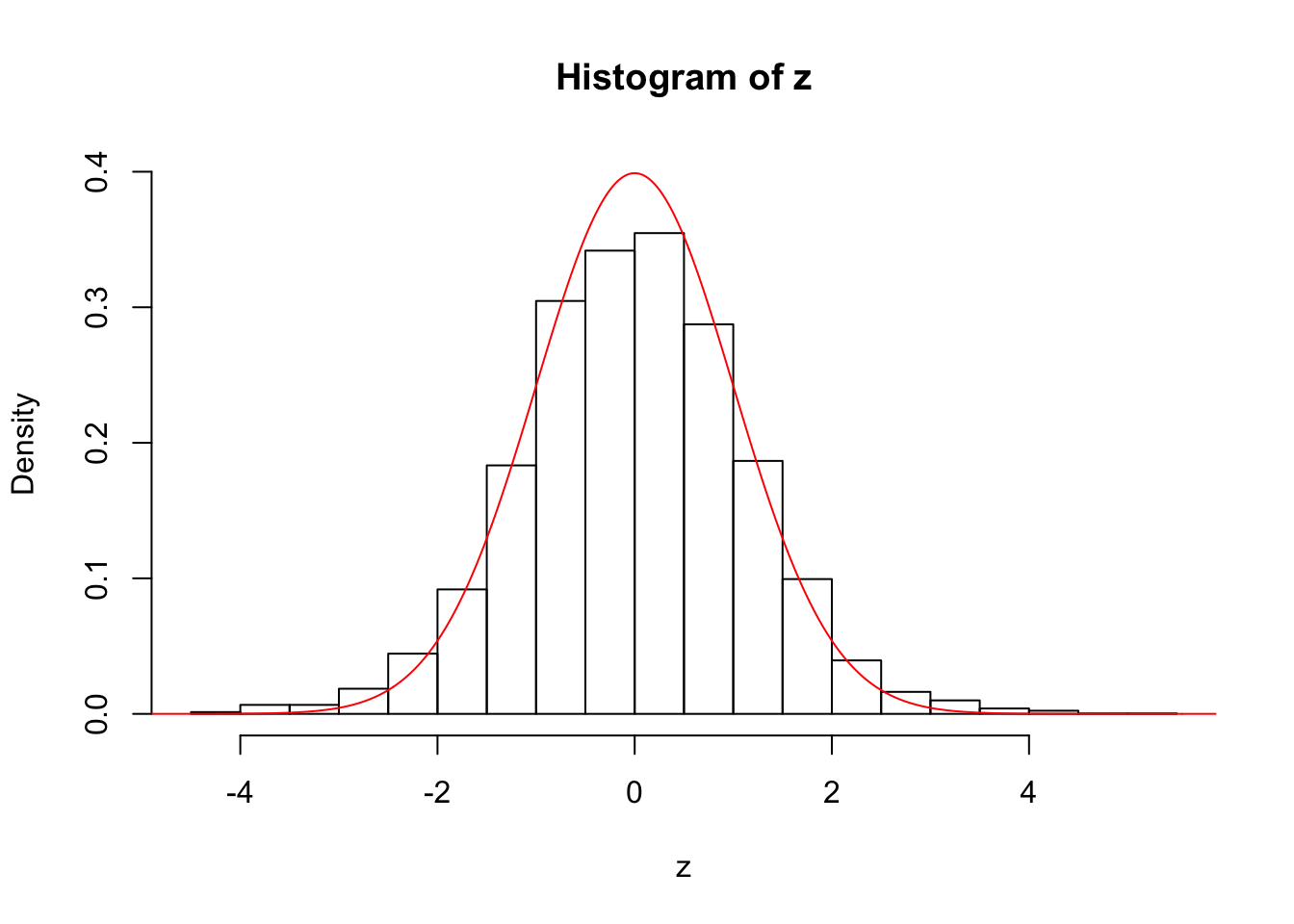
fit <- gdash(betahat, sebetahat, gd.ord = 10)
rand.sign <- sample(c(rep(1, nrow(prostate) / 2), rep(-1, nrow(prostate) / 2)))
fit.sym <- gdash(rand.sign * betahat, sebetahat, gd.ord = 10)Warning in rand.sign * betahat: longer object length is not a multiple of
shorter object lengthfit.locfdr <- locfdr(z)
police <- scan("https://web.stanford.edu/~hastie/CASI_files/DATA/police.txt")
fit <- gdash(police, rep(1, length(police)), gd.ord = 10)Warning in REBayes::KWDual(A, rep(1, k), normalize(w), control = control): estimated mixing distribution has some negative values:
consider reducing rtol
Warning in REBayes::KWDual(A, rep(1, k), normalize(w), control = control): estimated mixing distribution has some negative values:
consider reducing rtol
Warning in REBayes::KWDual(A, rep(1, k), normalize(w), control = control): estimated mixing distribution has some negative values:
consider reducing rtolDTI <- scan("https://web.stanford.edu/~hastie/CASI_files/DATA/DTI.txt")Session information
sessionInfo()R version 3.4.3 (2017-11-30)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS High Sierra 10.13.4
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] lattice_0.20-35 scales_0.5.0 locfdr_1.1-8
[4] limma_3.34.4 ashr_2.2-2 Rmosek_8.0.69
[7] PolynomF_1.0-1 CVXR_0.95 REBayes_1.2
[10] Matrix_1.2-12 SQUAREM_2017.10-1 EQL_1.0-0
[13] ttutils_1.0-1
loaded via a namespace (and not attached):
[1] gmp_0.5-13.1 Rcpp_0.12.16 pillar_1.0.1
[4] plyr_1.8.4 compiler_3.4.3 git2r_0.21.0
[7] R.methodsS3_1.7.1 R.utils_2.6.0 iterators_1.0.9
[10] tools_3.4.3 digest_0.6.15 bit_1.1-12
[13] tibble_1.4.1 gtable_0.2.0 evaluate_0.10.1
[16] rlang_0.1.6 foreach_1.4.4 yaml_2.1.18
[19] parallel_3.4.3 Rmpfr_0.6-1 ECOSolveR_0.4
[22] stringr_1.3.0 knitr_1.20 rprojroot_1.3-2
[25] bit64_0.9-7 grid_3.4.3 qvalue_2.10.0
[28] R6_2.2.2 rmarkdown_1.9 reshape2_1.4.3
[31] ggplot2_2.2.1 magrittr_1.5 splines_3.4.3
[34] MASS_7.3-47 backports_1.1.2 codetools_0.2-15
[37] htmltools_0.3.6 scs_1.1-1 assertthat_0.2.0
[40] colorspace_1.3-2 stringi_1.1.6 lazyeval_0.2.1
[43] munsell_0.4.3 doParallel_1.0.11 pscl_1.5.2
[46] truncnorm_1.0-7 R.oo_1.21.0 This R Markdown site was created with workflowr