FDR / \(s\) value on Correlated Null: betahat = betahat, sebetahat = betahat / zscore
Lei Sun
2017-02-16
Last updated: 2017-02-16
Code version: ca57b48
Introduction
Apply two FDR-controlling procedures, Benjamini–Hochberg 1995 (“BH”) and Benjamini-Yekutieli 2001 (“BY”), and two \(s\) value models, ash and truncash (with the threshold \(T = 1.96\)) to the simulated, correlated null data. The data are obtained from 5 vs 5 GTEx/Liver samples and 10K top expressed genes, and \(1000\) independent simulation trials.
Compare the numbers of false discoveries (by definition, all discoveries should be false) obtained by these four methods, using FDR \(\leq 0.05\) and \(s\)-value \(\leq 0.05\) as cutoffs.
Simulation: \(p\) values for BH and BY procedures; \(\hat\beta = \hat\beta\), \(\hat s = \hat\beta / \hat z\), for ash and truncash.
\(\hat\beta\) obtained from the limma::lmFit step of the pipeline, \(\hat z\) obtained from the last step of the pipeline.
library(ashr)
source("../code/truncash.R")p = read.table("../output/p_null_liver_777.txt")
z = read.table("../output/z_null_liver_777.txt")
betahat = read.table("../output/betahat_null_liver_777.txt")
m = dim(p)[1]
n = dim(p)[2]
fd.bh = fd.by = fd.ash = fd.truncash = c()
for (i in 1:m) {
p_BH = p.adjust(p[i, ], method = "BH")
fd.bh[i] = sum(p_BH <= 0.05)
p_BY = p.adjust(p[i, ], method = "BY")
fd.by[i] = sum(p_BY <= 0.05)
betahat_trial = as.numeric(betahat[i, ])
sebetahat_trial = - betahat_trial / as.numeric(z[i, ])
fit.ash = ashr::ash(betahat_trial, sebetahat_trial, method = "fdr", mixcompdist = "normal")
fd.ash[i] = sum(ashr::get_svalue(fit.ash) <= 0.05)
fit.truncash = truncash(betahat_trial, sebetahat_trial, t = qnorm(0.975))
fd.truncash[i] = sum(get_svalue(fit.truncash) <= 0.05)
}Result
Simulated under the global null, FWER \(=\) FDR.
Estimated FWER or FDR by BH
fdr.bh = mean(fd.bh >= 1)
fdr.bh[1] 0.046Estimated FWER or FDR by BY
fdr.by = mean(fd.by >= 1)
fdr.by[1] 0.006Estimated FWER or FDR by ash
fdr.ash = mean(fd.ash >= 1)
fdr.ash[1] 0.096Estimated FWER or FDR by truncash
fdr.truncash = mean(fd.truncash >= 1)
fdr.truncash[1] 0.062Happenstance of false discoveries by four approaches
maxcount = max(c(fd.bh, fd.by, fd.ash, fd.truncash))
xlim = c(0, maxcount)
maxfreq = max(c(max(table(fd.bh)), max(table(fd.by)), max(table(fd.ash)), max(table(fd.truncash))))
ylim = c(0, maxfreq)
plot(table(fd.bh), xlab = "Number of False Discoveries / 10K", ylab = "Frequency", main = "Benjamini - Hochberg 1995", xlim = xlim, ylim = ylim)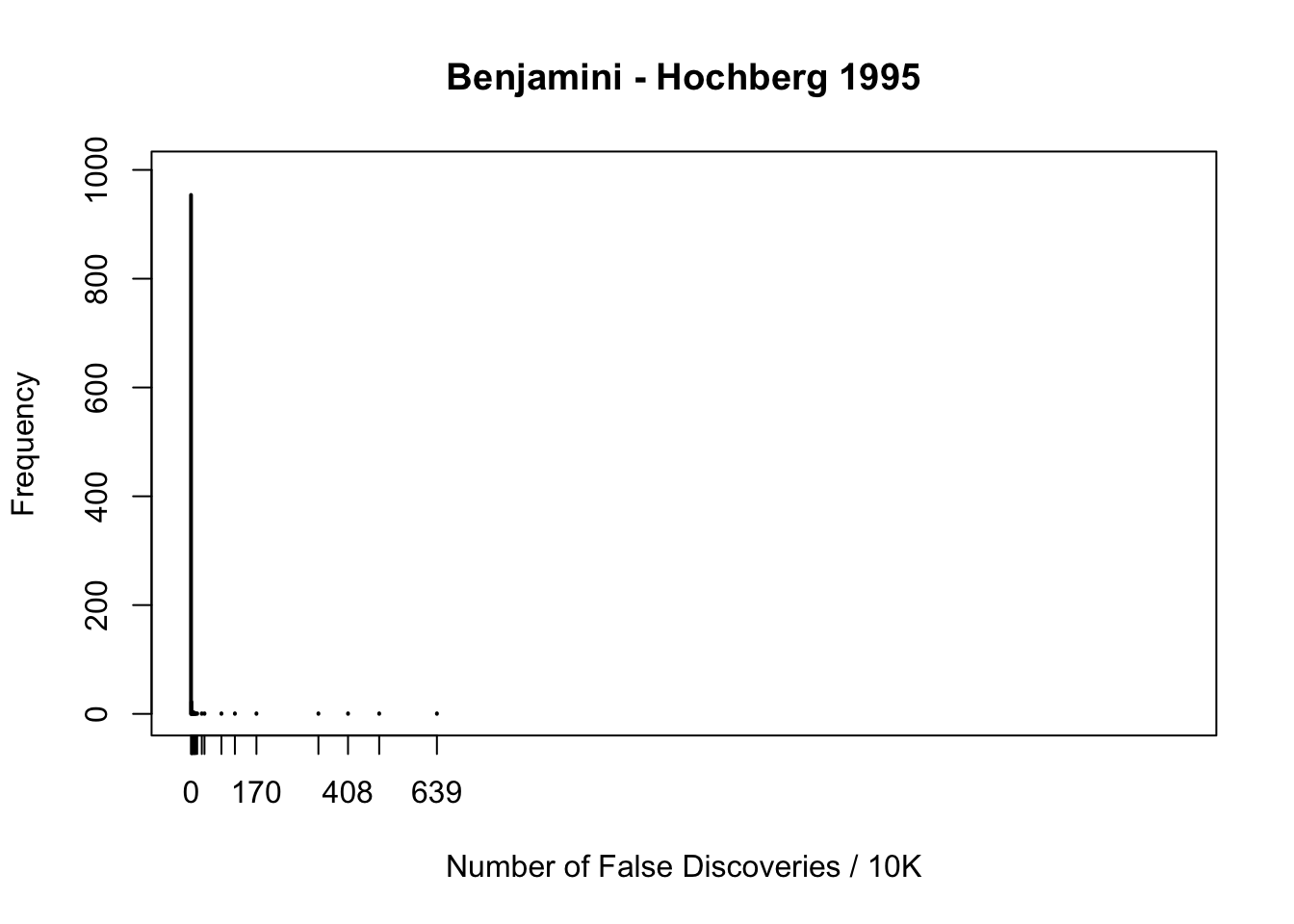
plot(table(fd.by), xlab = "Number of False Discoveries / 10K", ylab = "Frequency", main = "Benjamini - Yekutieli 2001", xlim = xlim, ylim = ylim)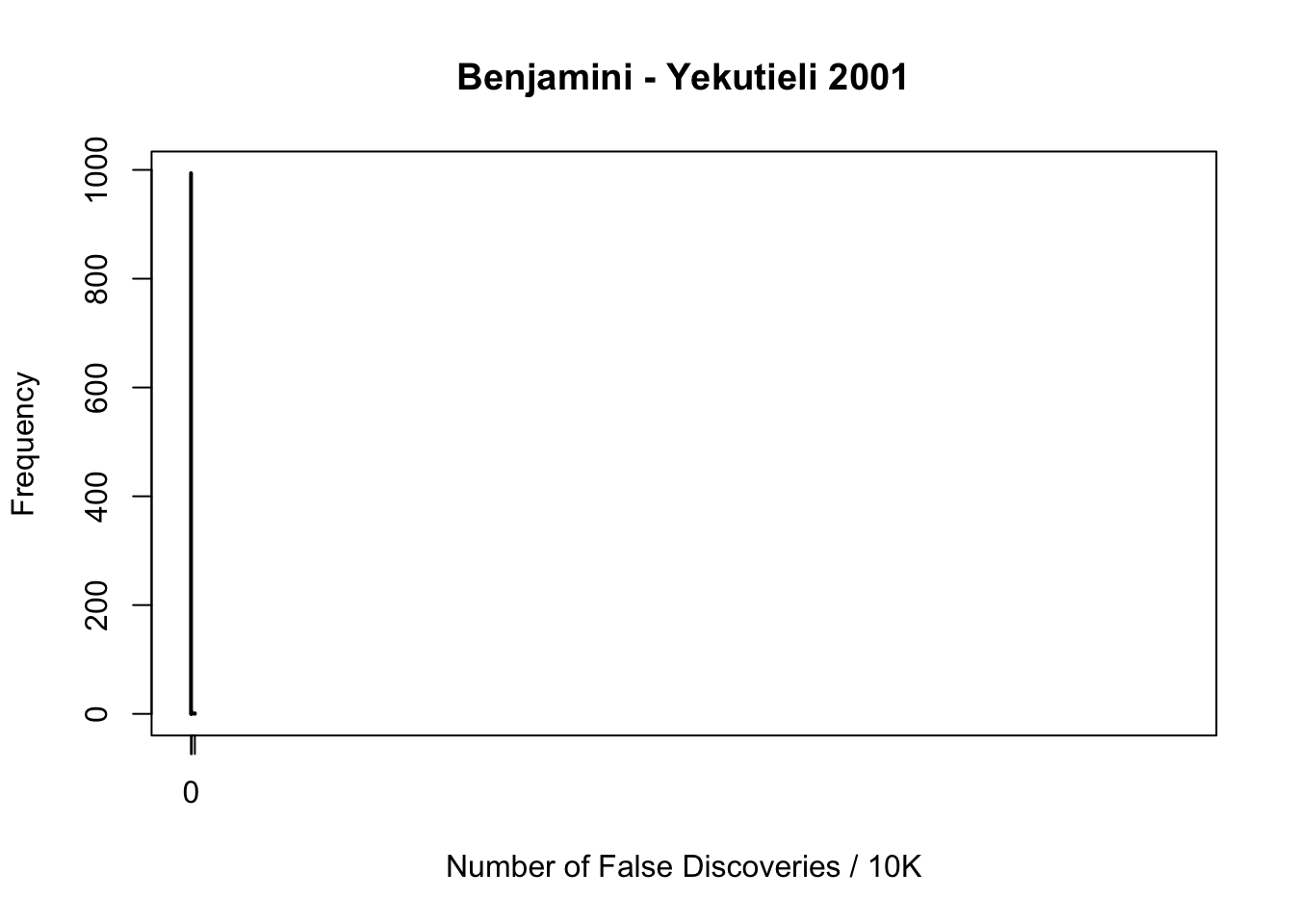
plot(table(fd.ash), xlab = "Number of False Discoveries / 10K", ylab = "Frequency", main = "ash", xlim = xlim, ylim = ylim)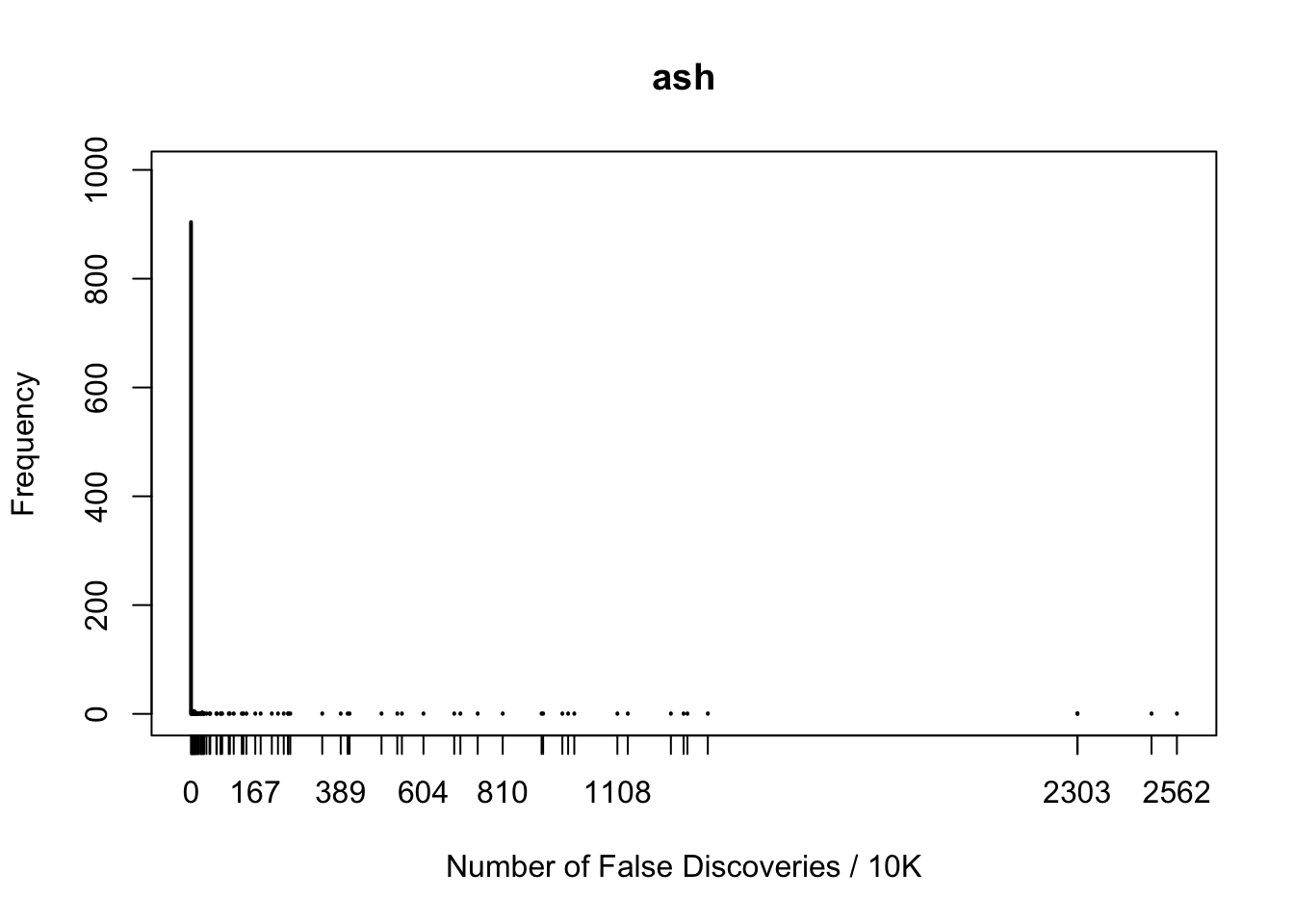
plot(table(fd.truncash), xlab = "Number of False Discoveries / 10K", ylab = "Frequency", main = "truncash", xlim = xlim, ylim = ylim)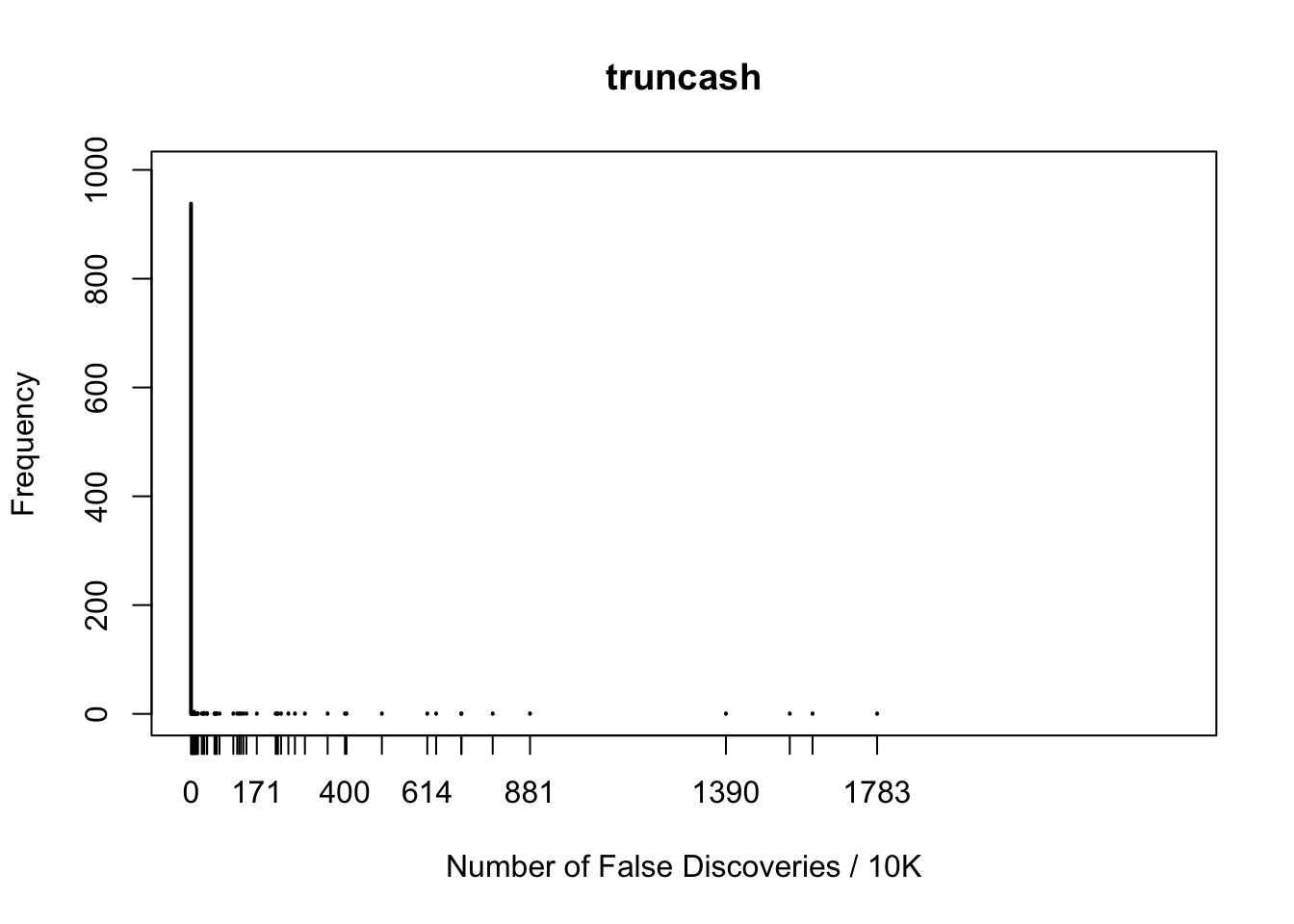
Comparison of the numbers of false discoveries by four approaches
m = length(fd.bh)
fd.ind = (1:m)[!((fd.bh == 0) & (fd.by == 0) & (fd.ash == 0) & (fd.truncash == 0))]
plot(1:length(fd.ind), fd.bh[fd.ind], pch = 4, ylim = xlim, xlab = "Trials with False Discoveries", ylab = "Number of False Discoveries / 10K")
points(1:length(fd.ind), fd.by[fd.ind], pch = 4, col = 2)
points(1:length(fd.ind), fd.ash[fd.ind], pch = 4, col = 3)
points(1:length(fd.ind), fd.truncash[fd.ind], pch = 4, col = 4)
legend("topright", c("BH", "BY", "ash", "truncash"), col = 1:4, pch = 4)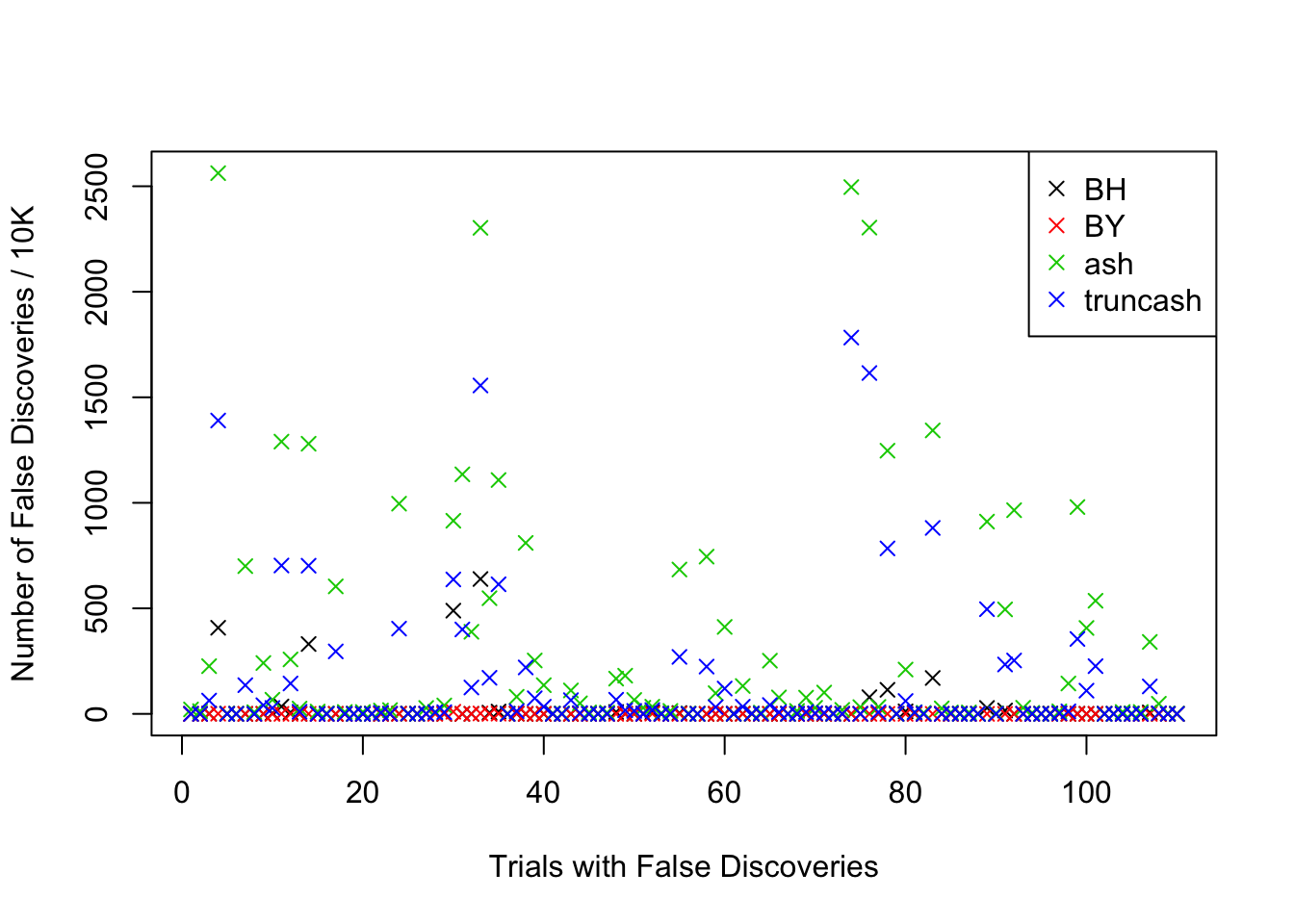
Session Information
sessionInfo()R version 3.3.2 (2016-10-31)
Platform: x86_64-apple-darwin13.4.0 (64-bit)
Running under: macOS Sierra 10.12.3
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] SQUAREM_2016.10-1 ashr_2.1.2
loaded via a namespace (and not attached):
[1] Rcpp_0.12.9 knitr_1.15.1 magrittr_1.5
[4] workflowr_0.3.0 MASS_7.3-45 doParallel_1.0.10
[7] pscl_1.4.9 lattice_0.20-34 foreach_1.4.3
[10] stringr_1.1.0 tools_3.3.2 parallel_3.3.2
[13] grid_3.3.2 git2r_0.18.0 htmltools_0.3.5
[16] iterators_1.0.8 yaml_2.1.14 rprojroot_1.2
[19] digest_0.6.9 codetools_0.2-15 evaluate_0.10
[22] rmarkdown_1.3 stringi_1.1.2 backports_1.0.5
[25] truncnorm_1.0-7 This R Markdown site was created with workflowr