plot_diagnostic in ashr
Lei Sun
2017-05-12
Last updated: 2017-05-12
Code version: fd54c84
library(ashr)set.seed(777)
x = rnorm(100)
qqnorm(x)
qqline(x)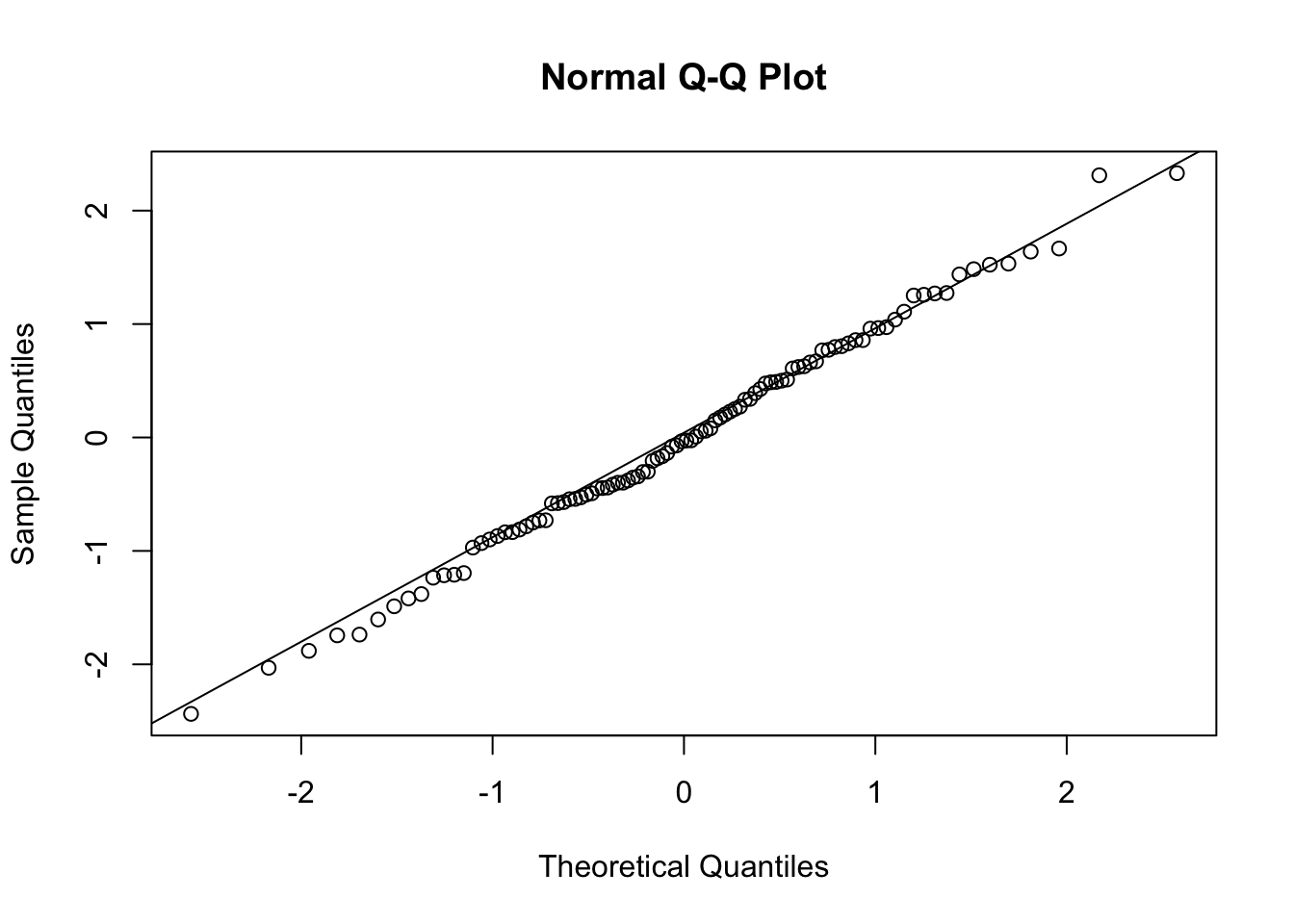
y = runif(100)
qqnorm(y)
qqline(y)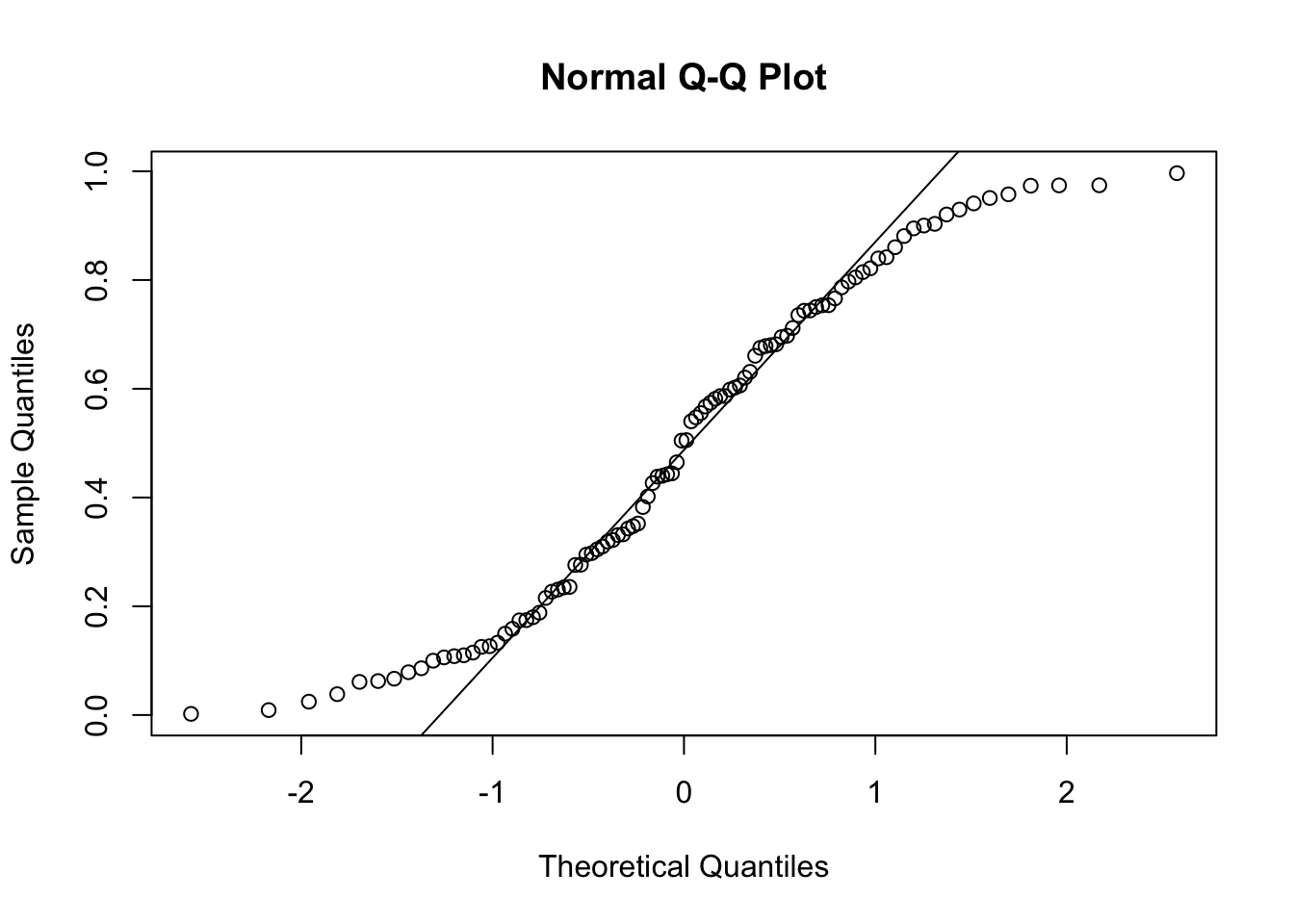
beta = runif(10000)
betahat = beta + rnorm(10000)
fit = ash(betahat, 1, mixcompdist = "normal", method = "fdr")
plot_diagnostic(fit)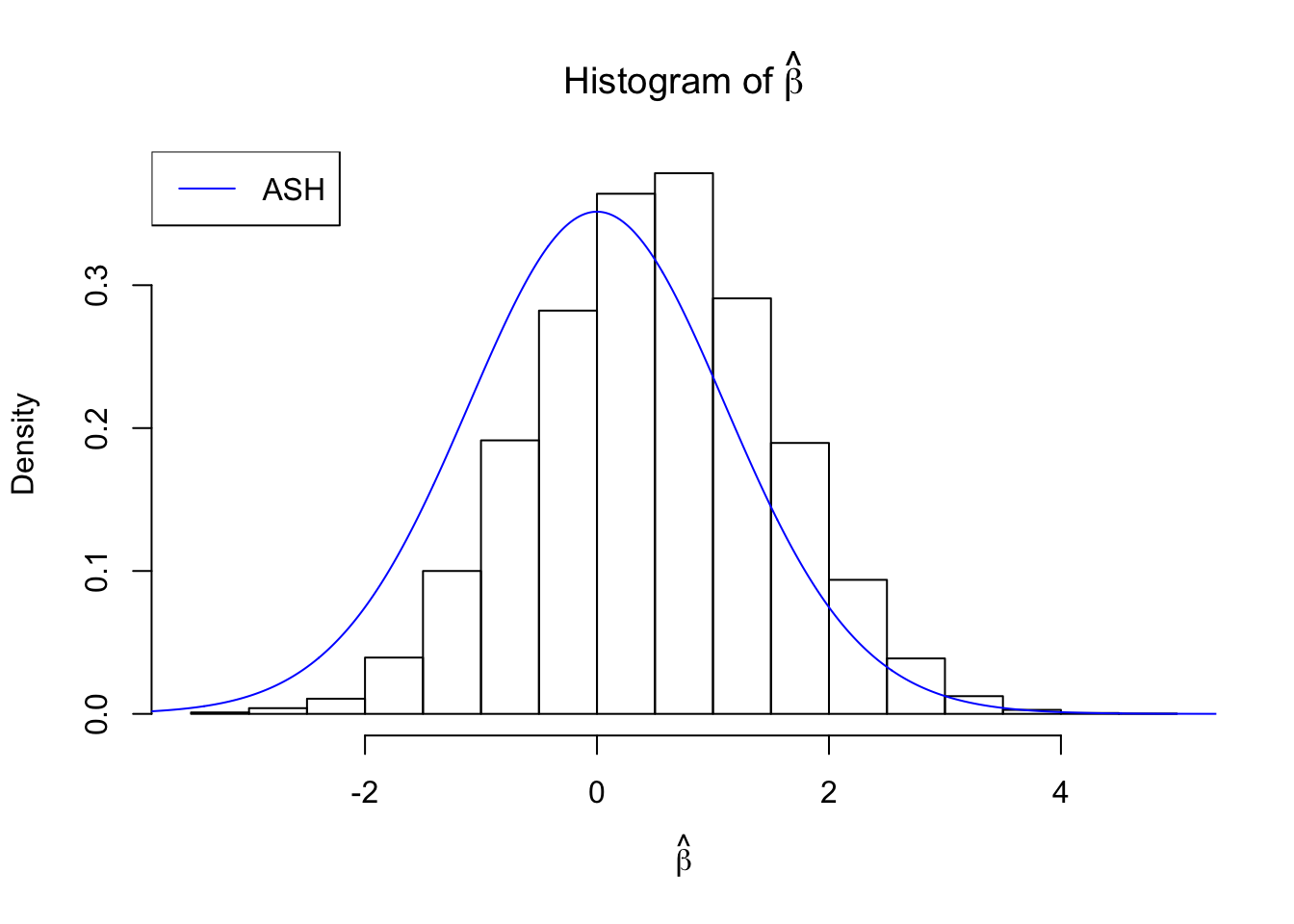
Press [enter] to see next plot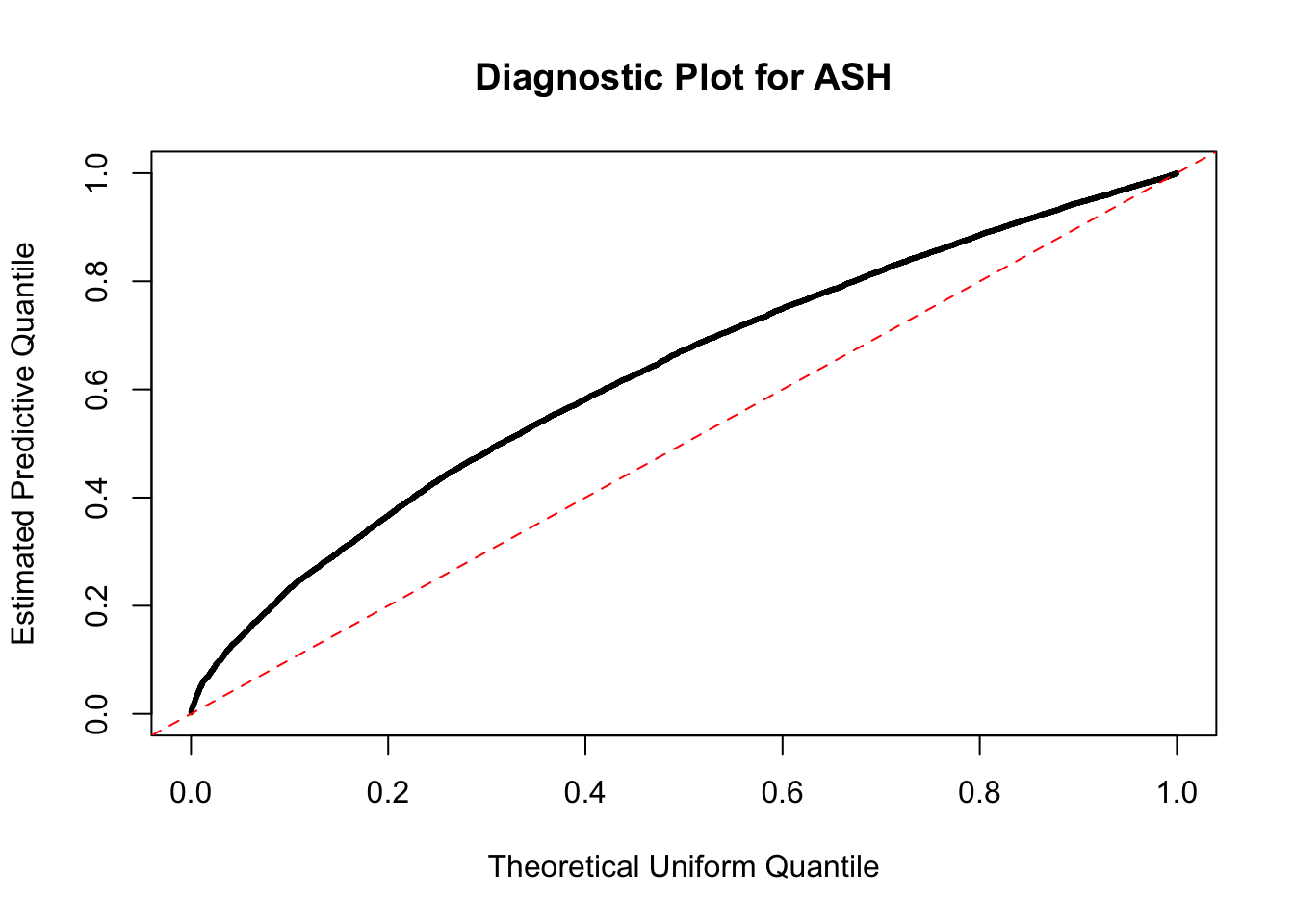
Press [enter] to see next plot
Press [enter] to see next plot
par(mfrow = c(2, 2))
fit = ash(rnorm(10000), 1, mixcompdist = "normal", method = "fdr")
plot_diagnostic(fit, breaks = 100)Press [enter] to see next plotPress [enter] to see next plotPress [enter] to see next plot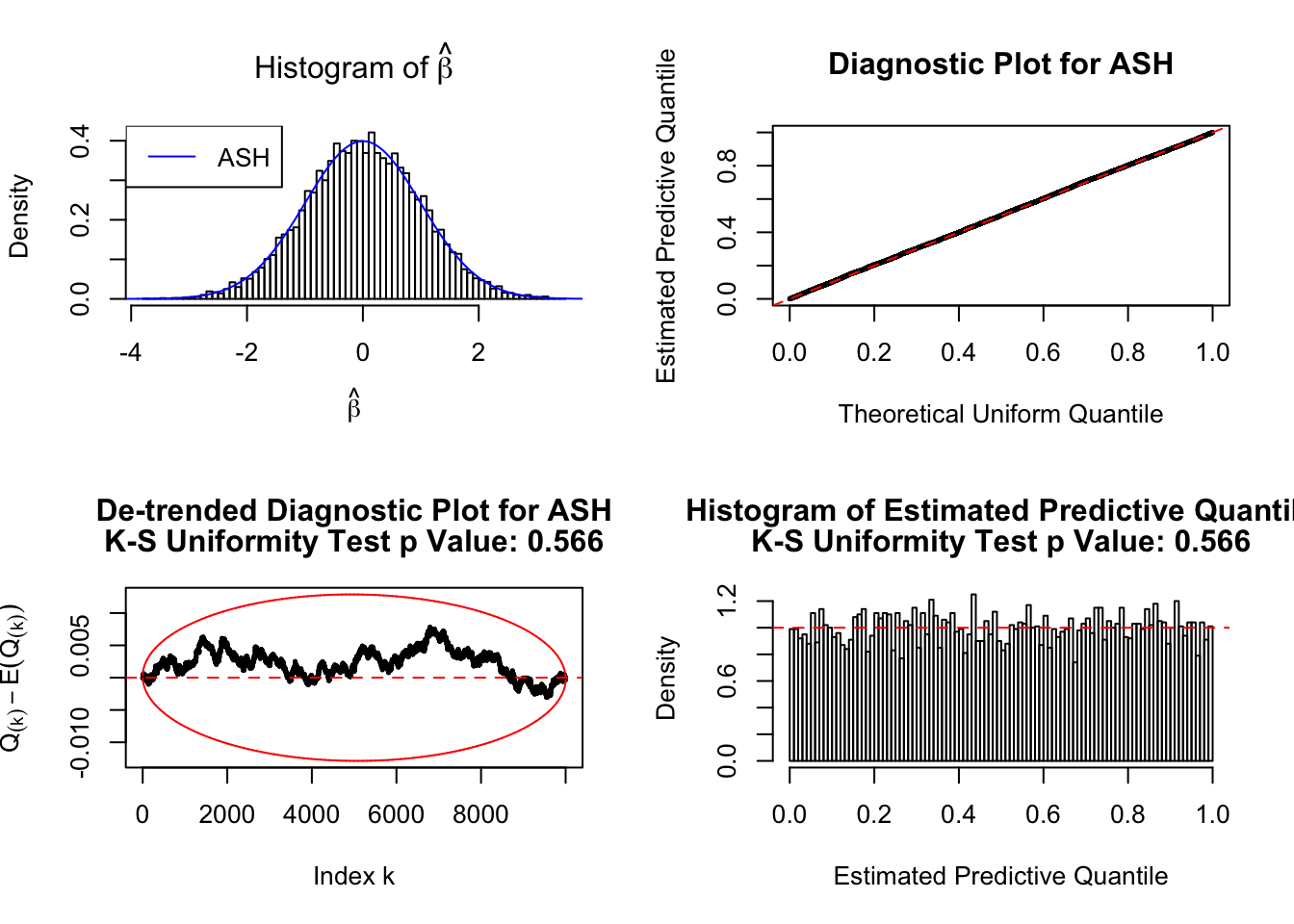
beta = sample(c(rnorm(5000, 1.5, 1), rnorm(5000, -1.5, 1)))
betahat = beta + rnorm(10000)
fit = ash(betahat, 1, mixcompdist = "normal", method = "fdr")
plot_diagnostic(fit, breaks = 100)Press [enter] to see next plotPress [enter] to see next plotPress [enter] to see next plot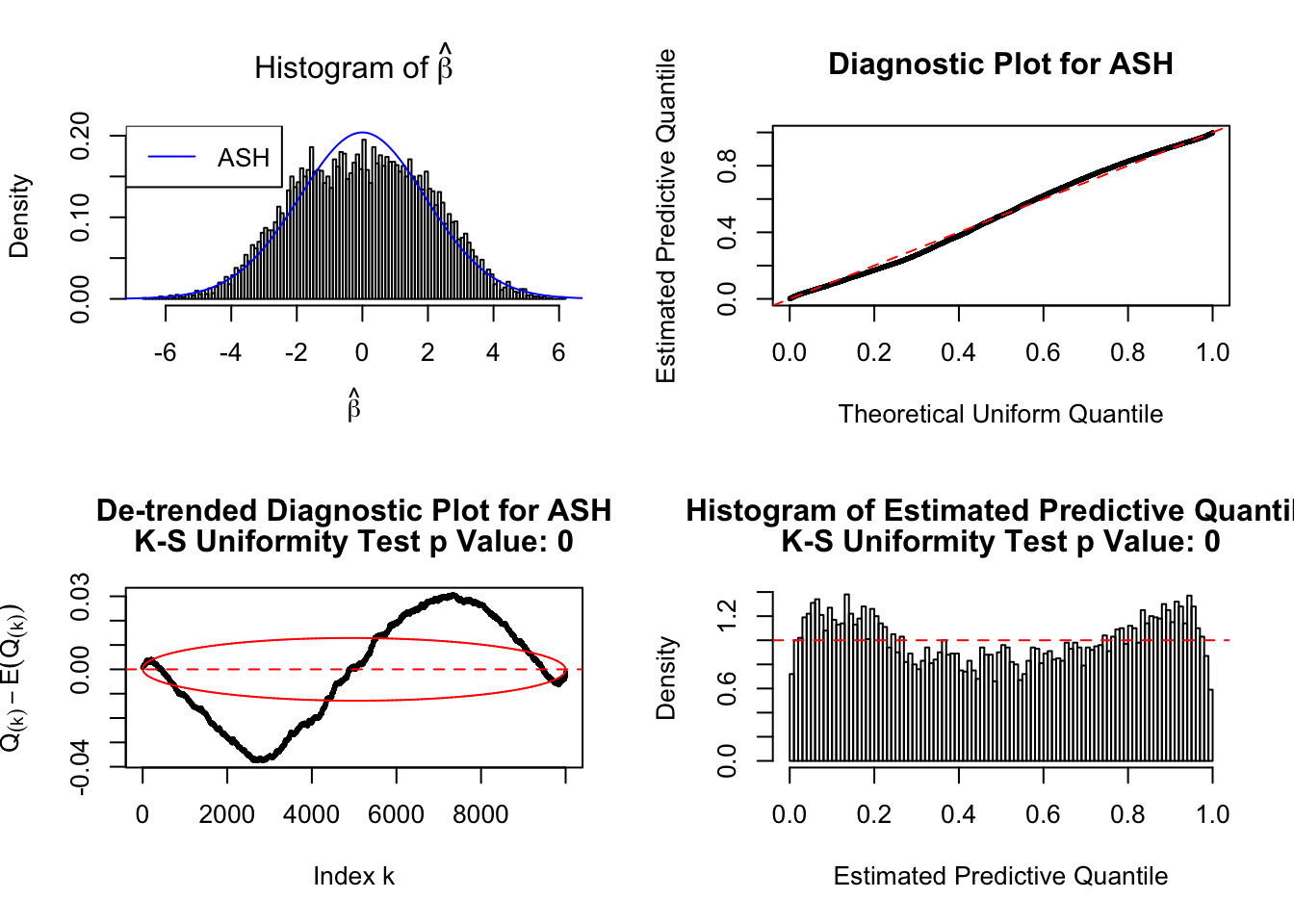
z = readRDS("../output/z_null_liver_777_select.RDS")
z = z$typical[[5]]
hist(z, breaks = 100, prob = TRUE, xlab = "Correlated N(0, 1) Z Scores", ylim = c(0, dnorm(0)), main = "Histogram of Correlated Z Scores")
lines(seq(-6, 6, 0.01), dnorm(seq(-6, 6, 0.01)), col = "red")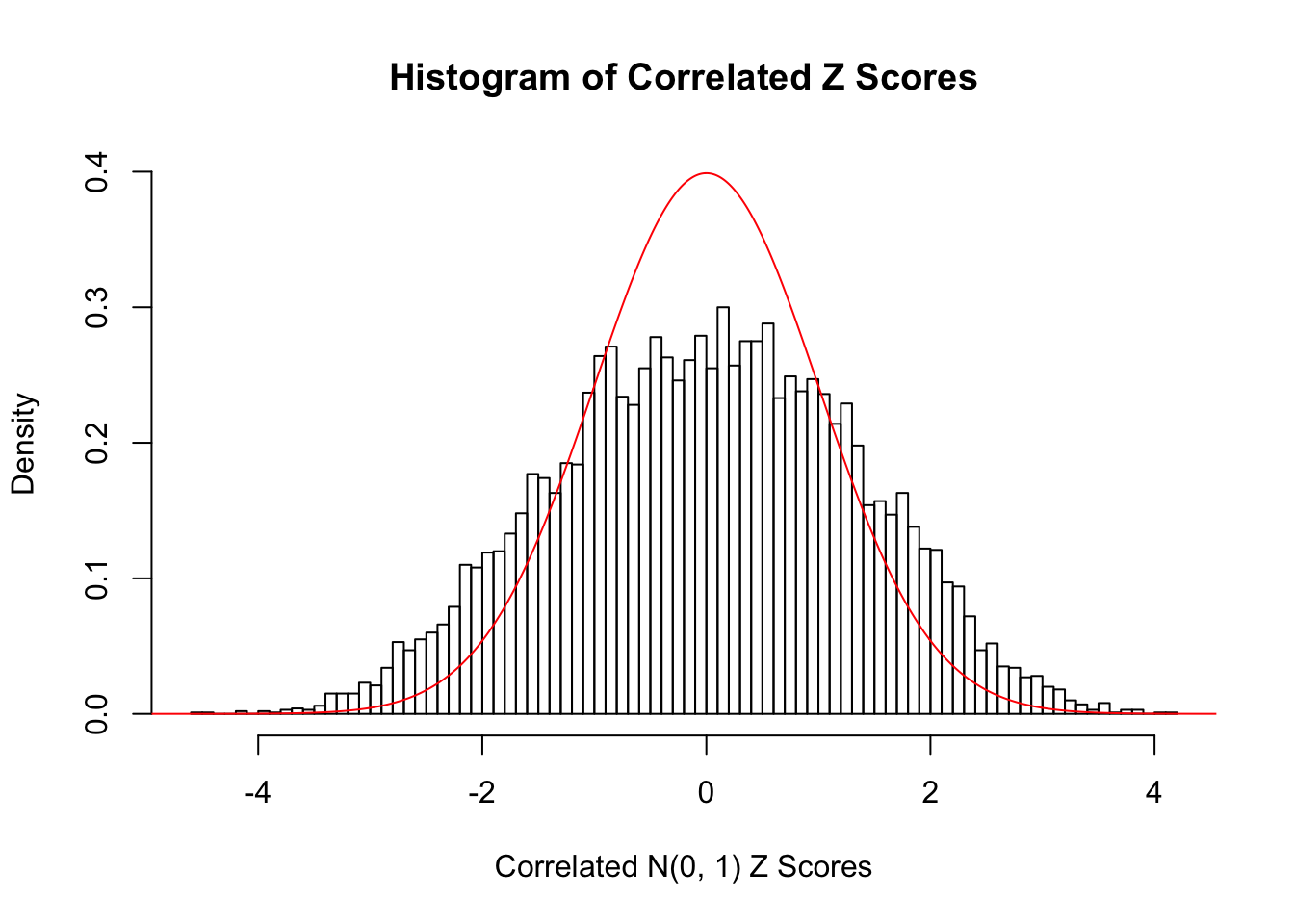
hist(z, breaks = 100, prob = TRUE, xlab = "Correlated N(0, 1) Z Scores", ylim = c(0, 0.1), main = "Histogram of Correlated Z Scores: Left Tail", xlim = c(-5, -2.5))
lines(seq(-6, 6, 0.01), dnorm(seq(-6, 6, 0.01)), col = "red")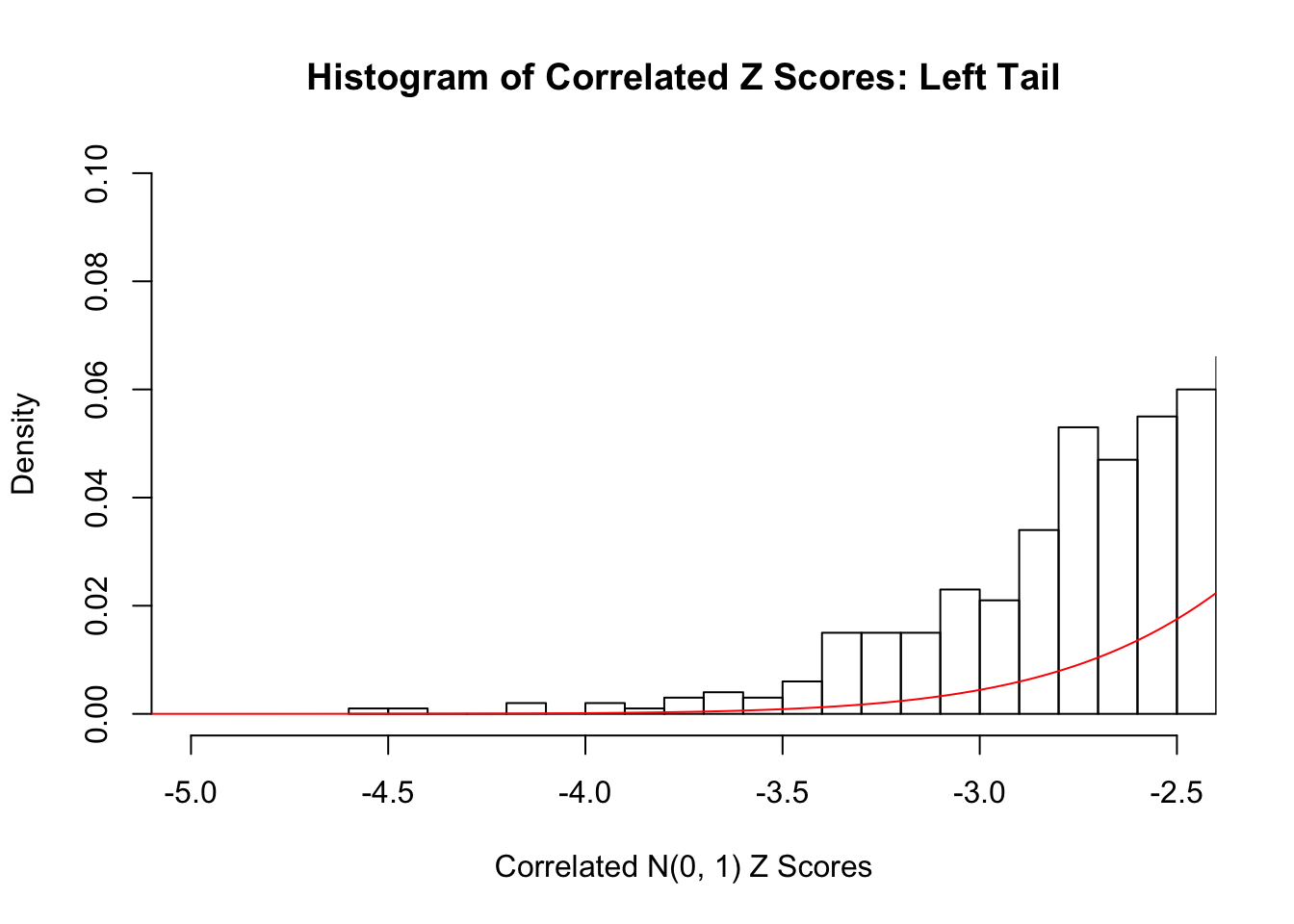
library(ashr)
fit = ashr::ash(z, 1, mixcompdist = "normal", method = "fdr")
plot_diagnostic(fit, breaks = 100)
Press [enter] to see next plot
Press [enter] to see next plot
Press [enter] to see next plot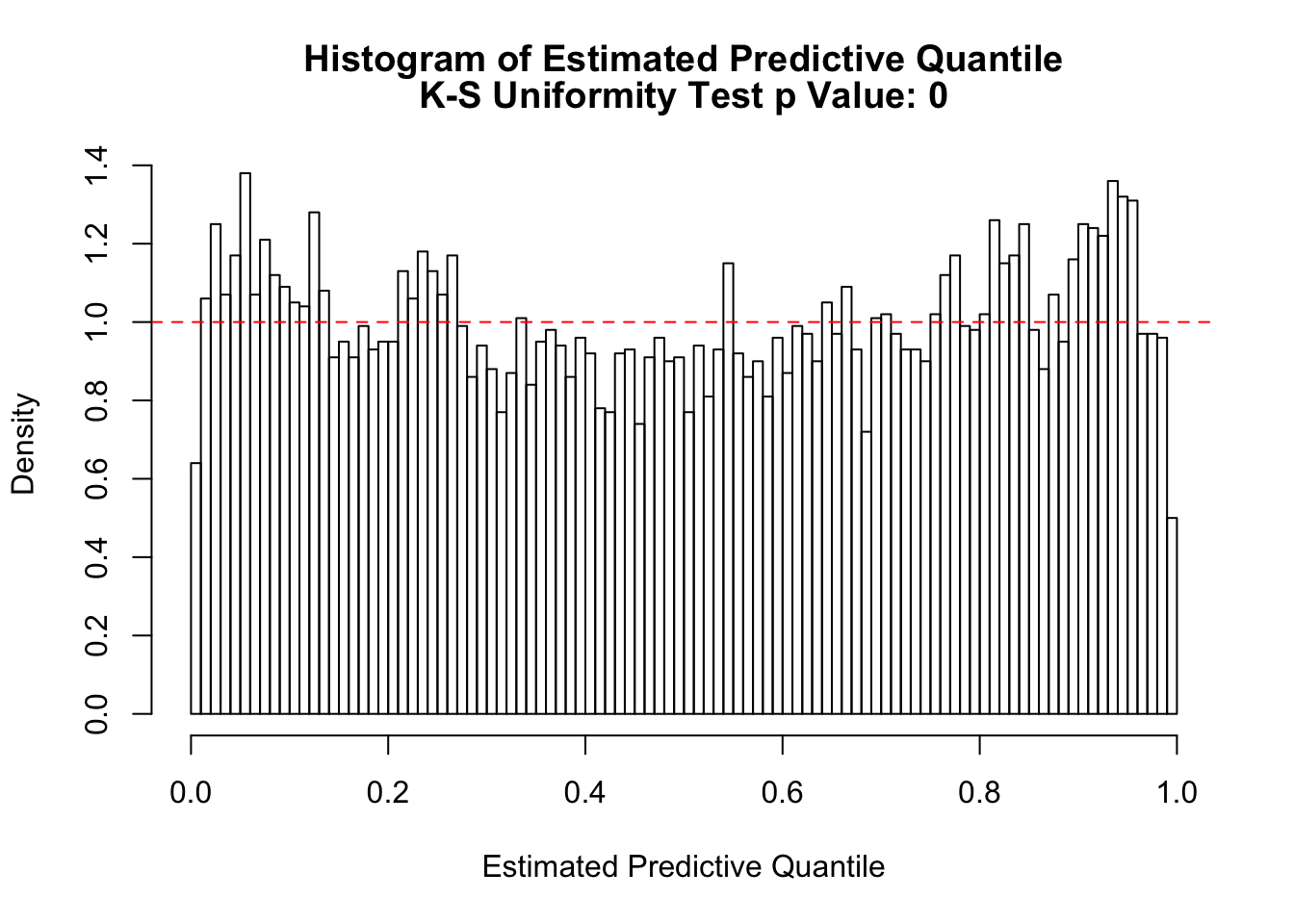
Session information
sessionInfo()R version 3.3.3 (2017-03-06)
Platform: x86_64-apple-darwin13.4.0 (64-bit)
Running under: macOS Sierra 10.12.4
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ashr_2.1-10
loaded via a namespace (and not attached):
[1] Rcpp_0.12.10 knitr_1.15.1 magrittr_1.5
[4] REBayes_0.62 MASS_7.3-45 doParallel_1.0.10
[7] pscl_1.4.9 SQUAREM_2016.10-1 lattice_0.20-34
[10] foreach_1.4.3 stringr_1.2.0 tools_3.3.3
[13] parallel_3.3.3 grid_3.3.3 git2r_0.18.0
[16] htmltools_0.3.5 iterators_1.0.8 assertthat_0.1
[19] yaml_2.1.14 rprojroot_1.2 digest_0.6.11
[22] Matrix_1.2-8 codetools_0.2-15 evaluate_0.10
[25] rmarkdown_1.3 stringi_1.1.2 Rmosek_7.1.3
[28] backports_1.0.5 truncnorm_1.0-7 This R Markdown site was created with workflowr