CASH Simulations
Lei Sun
2018-02-10
Last updated: 2018-04-27
Code version: 8c99563
\(\sigma_\beta = 2.5\), 4 Gaussian derivatives
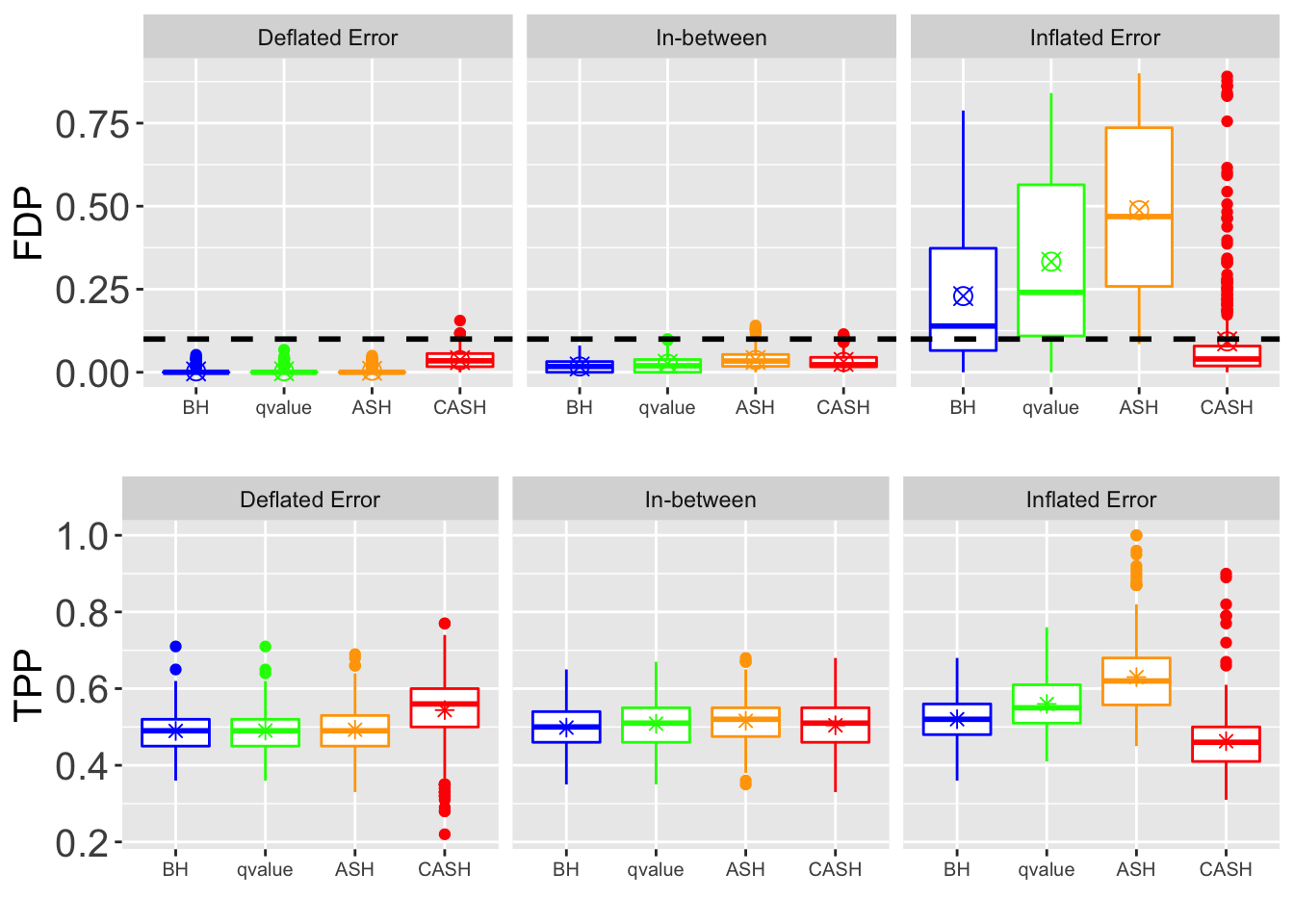
FDR, pFDR Calibration at each sparsity level
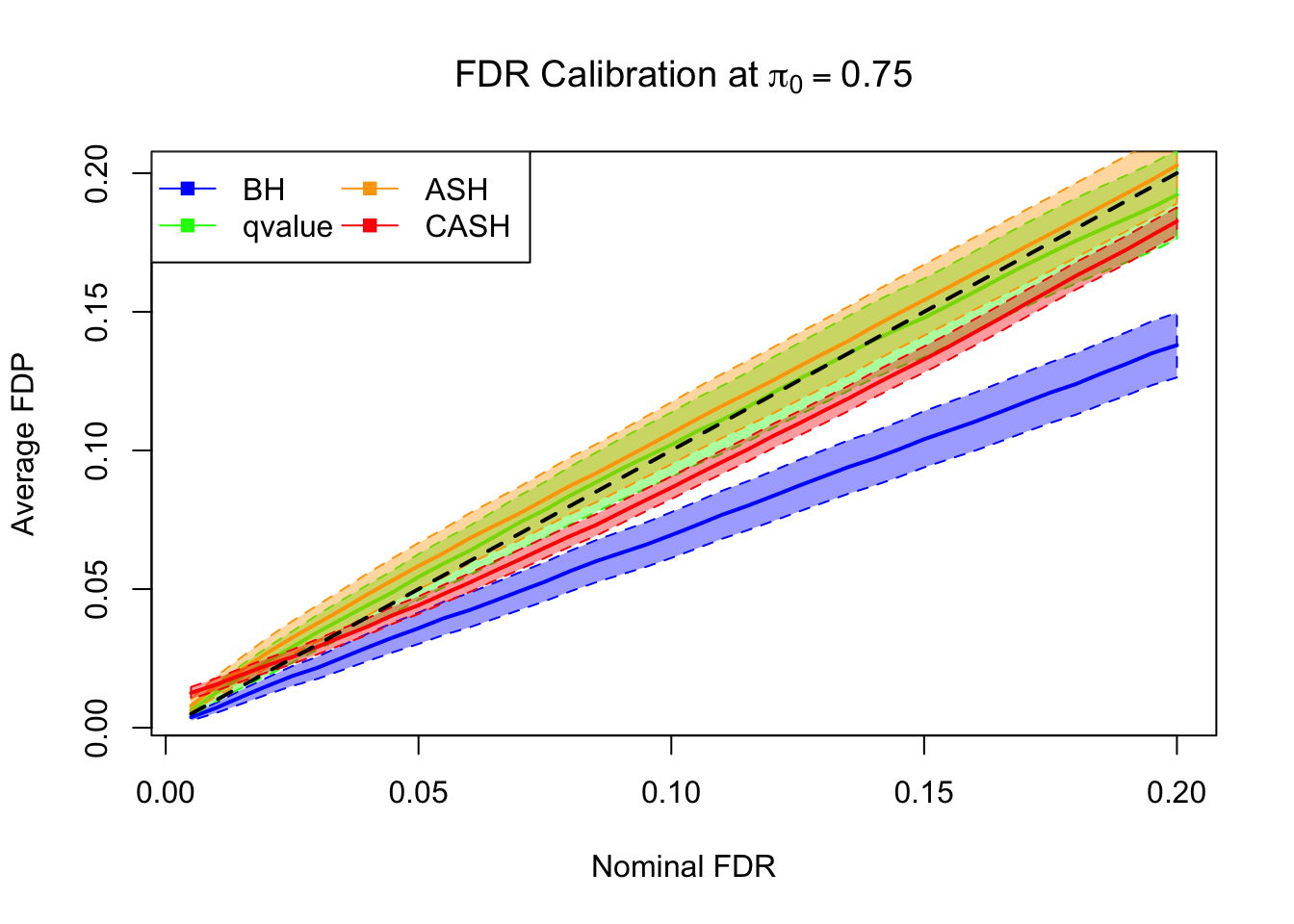
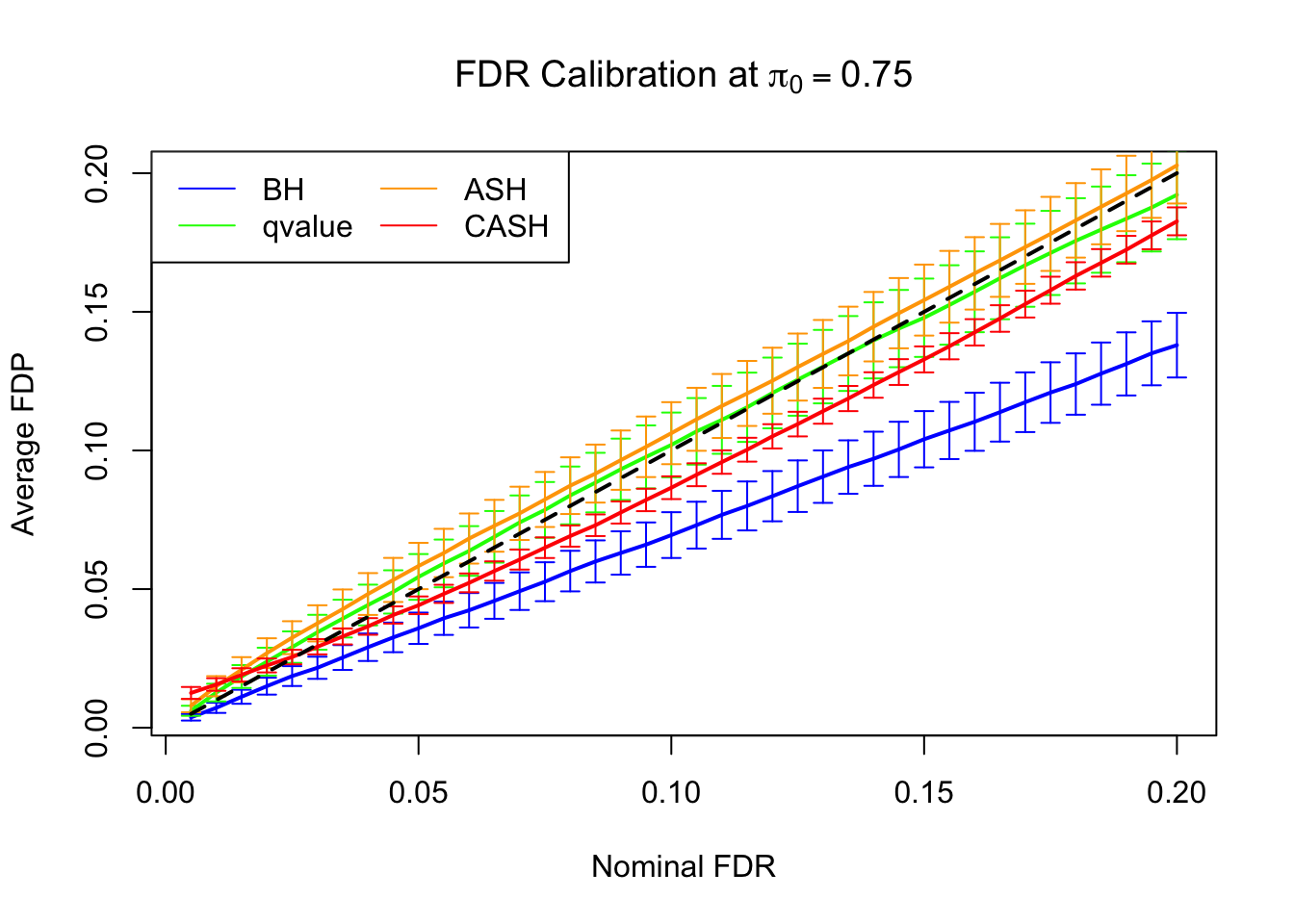
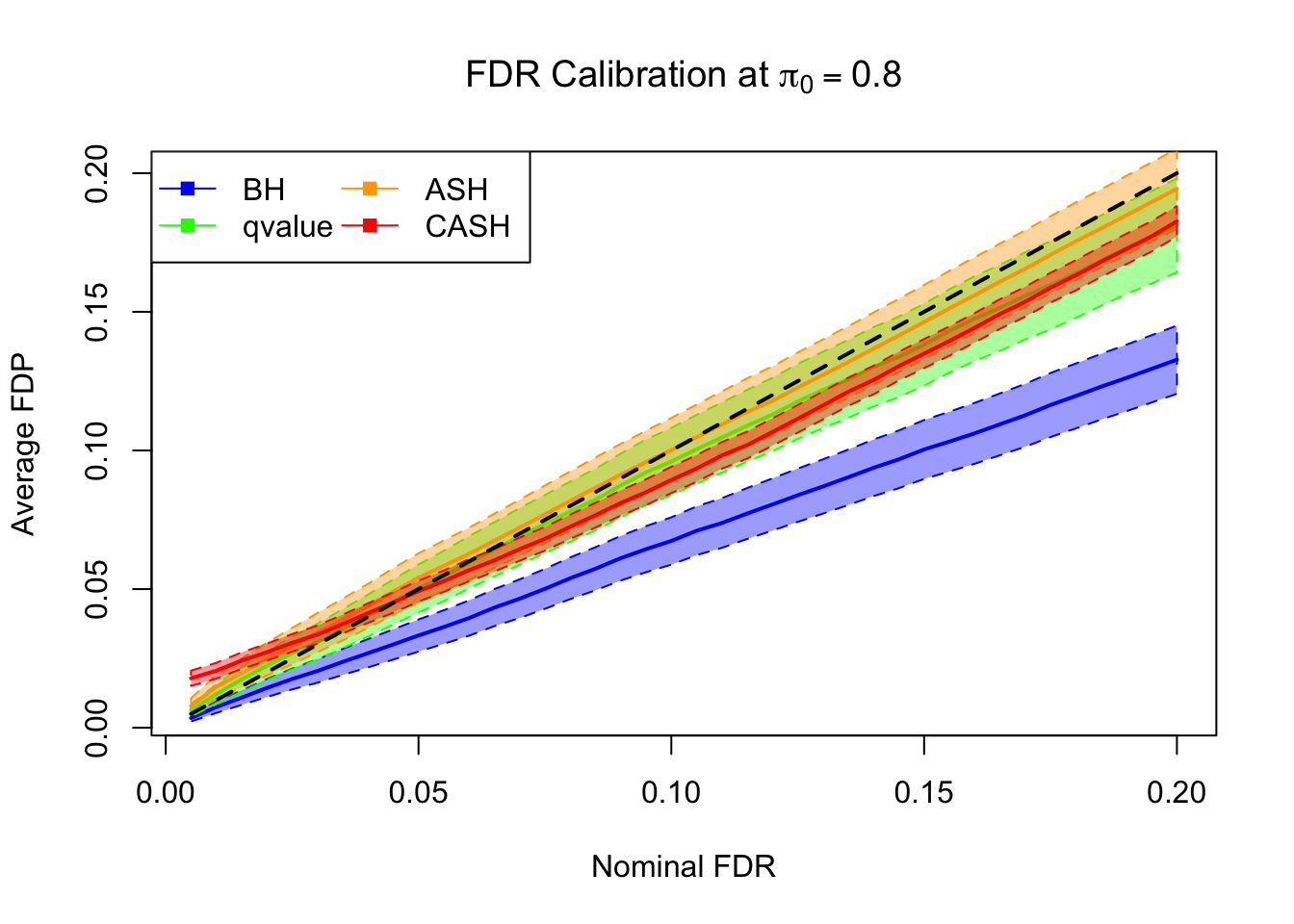
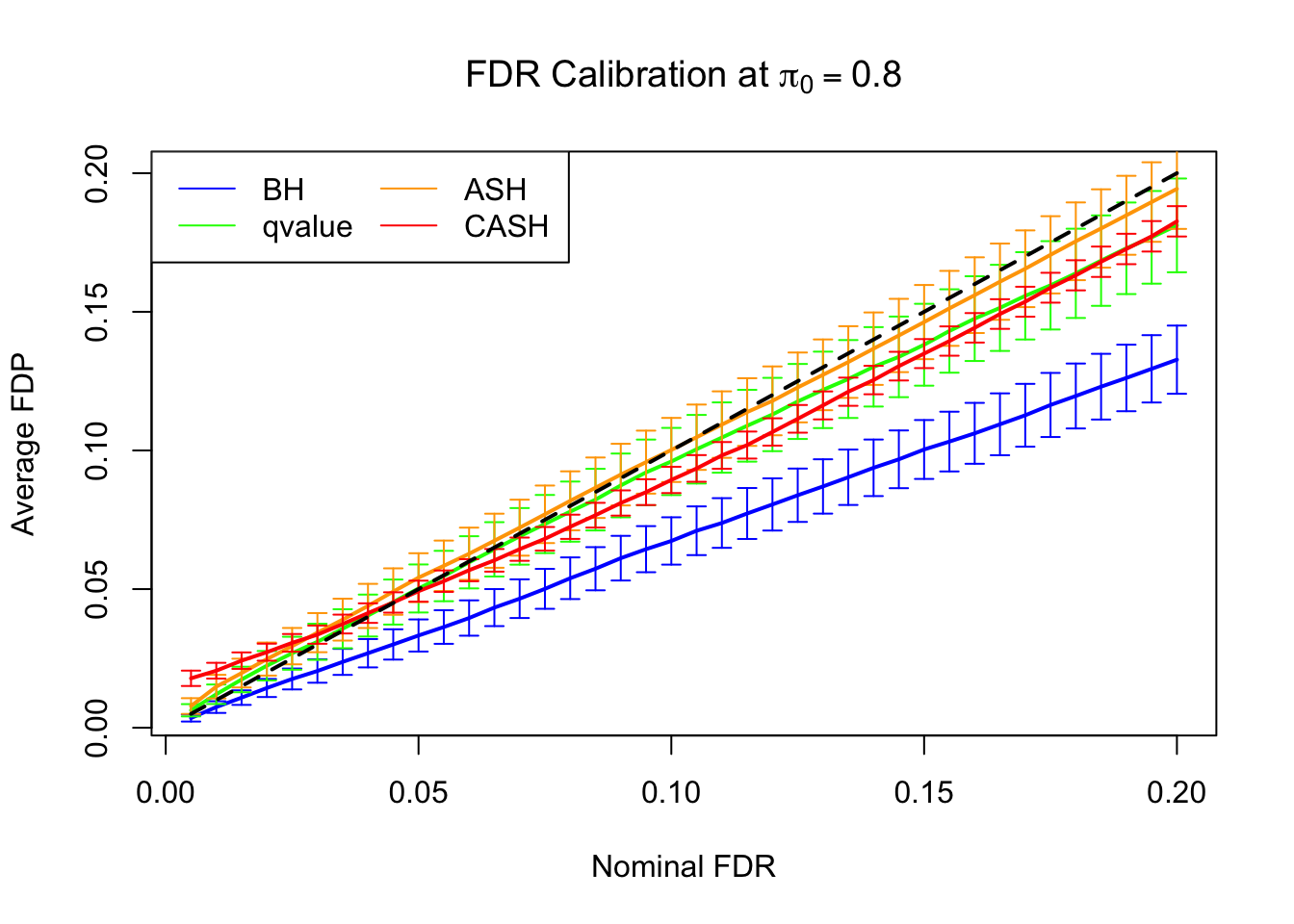
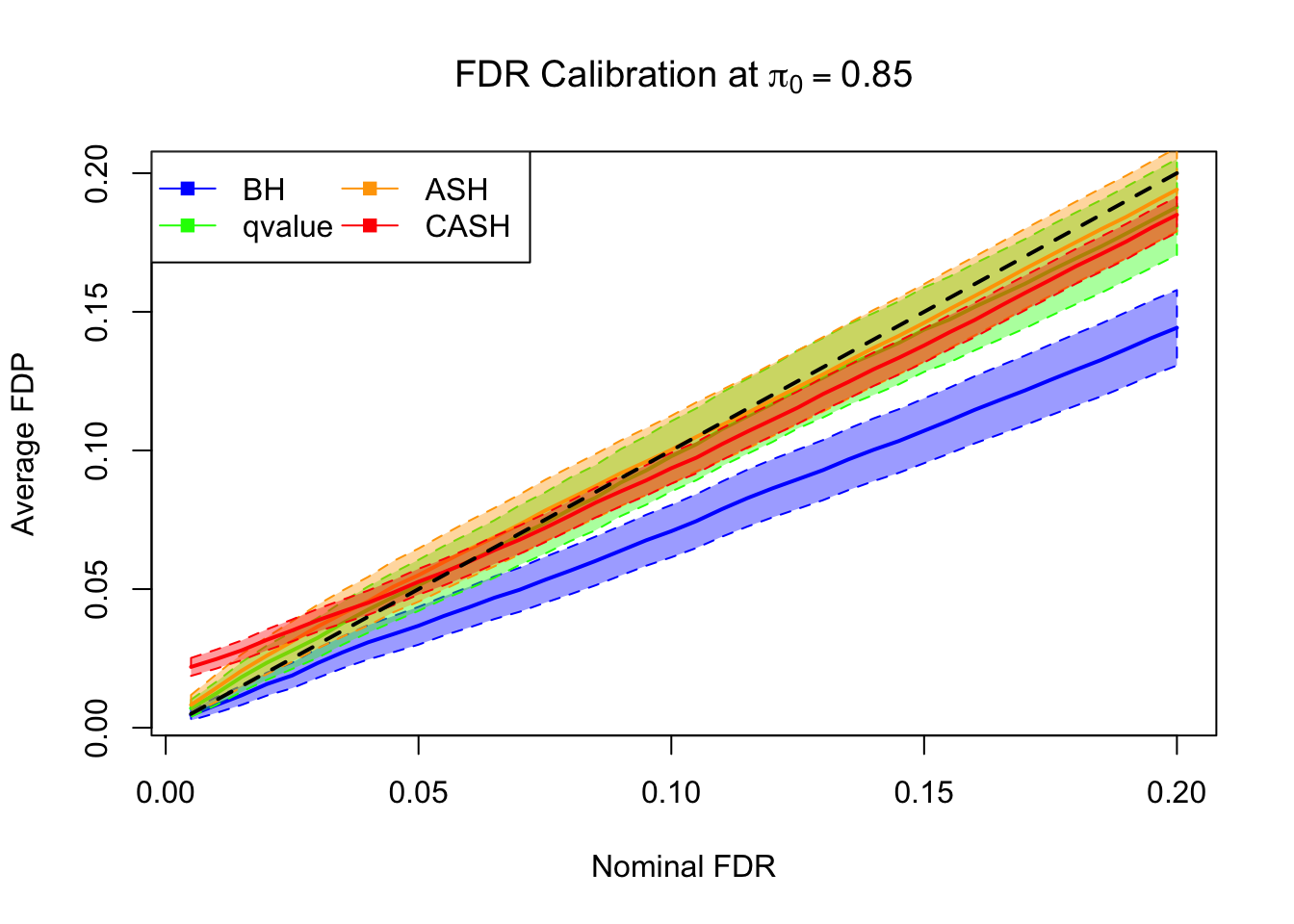
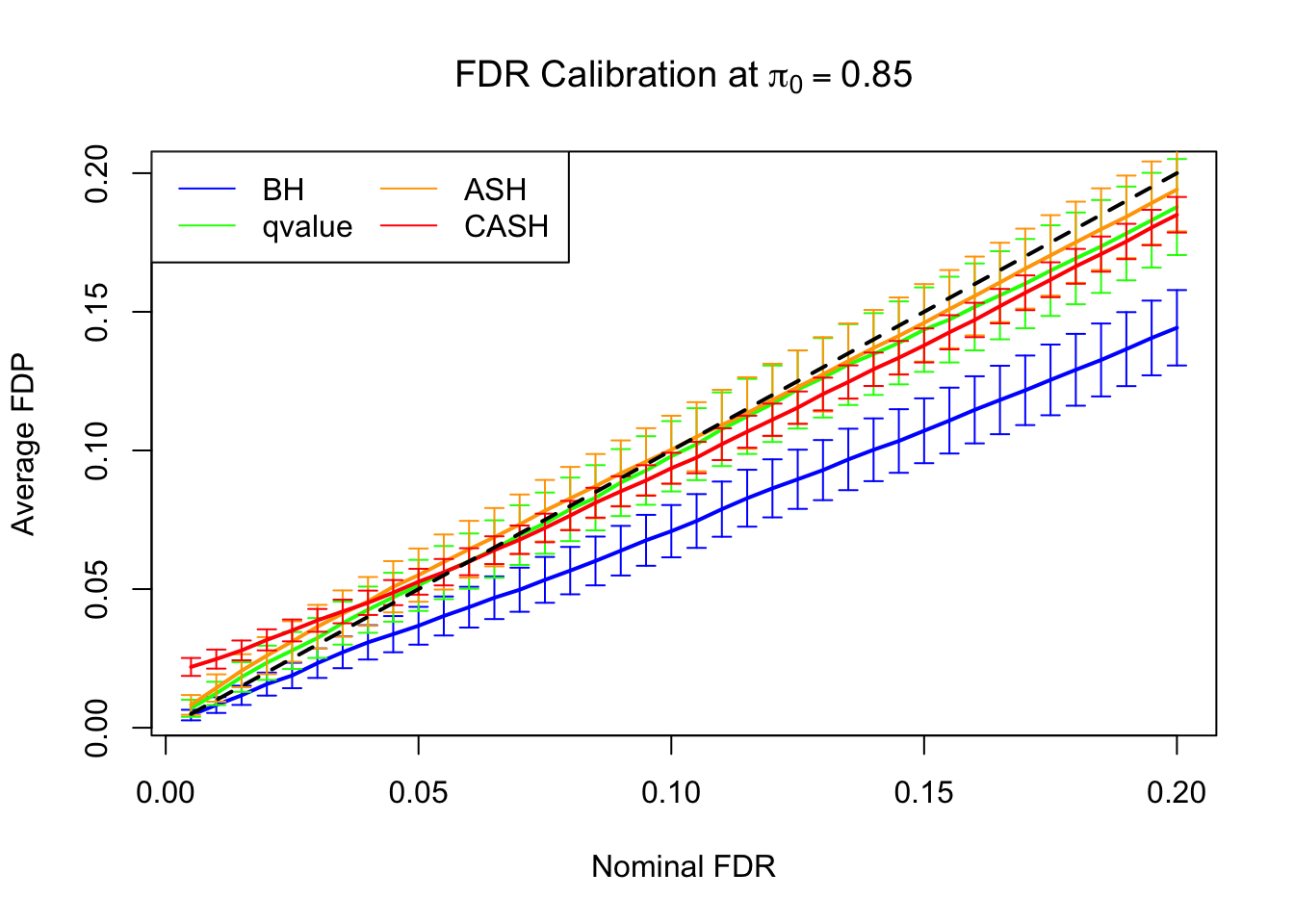
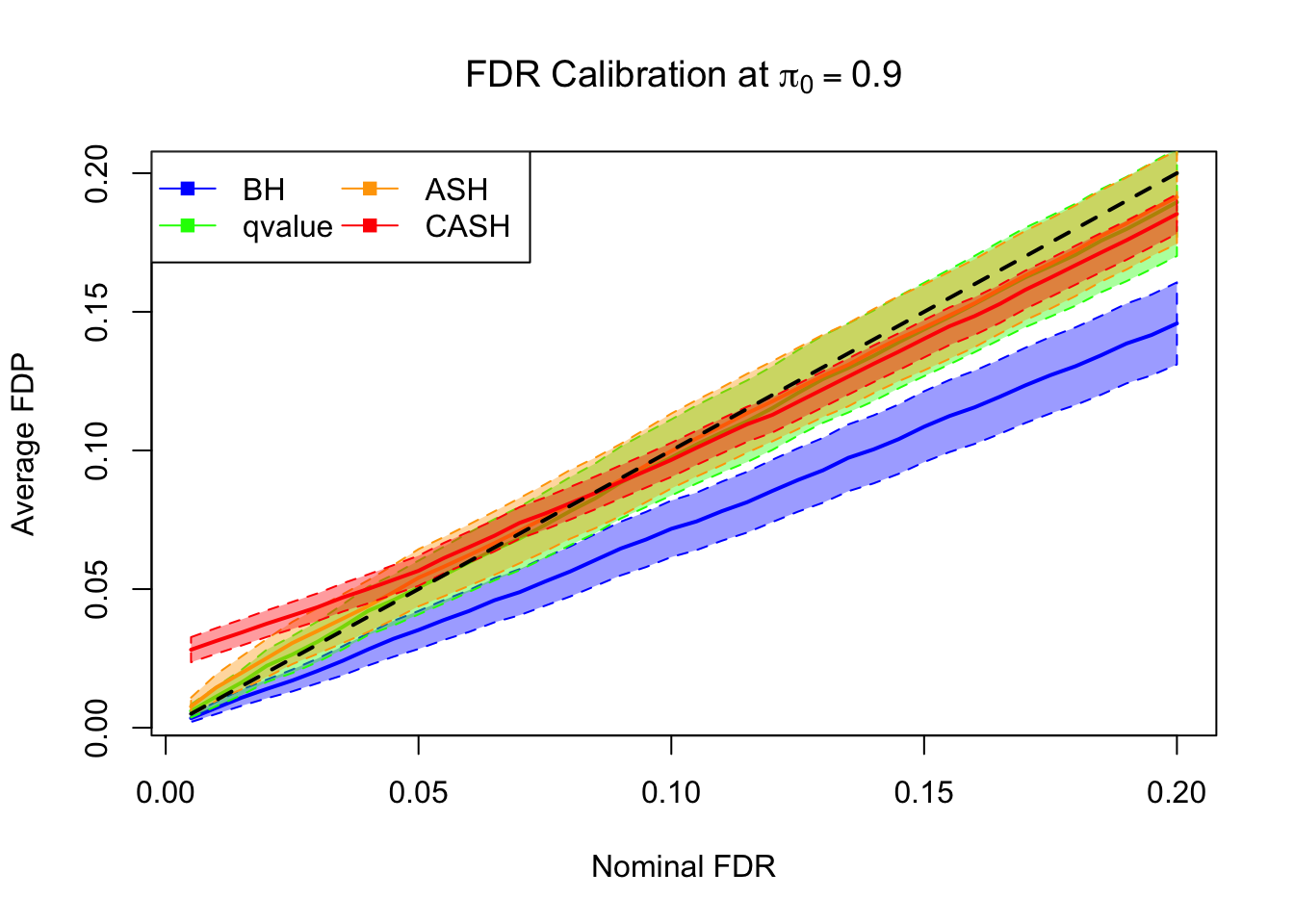
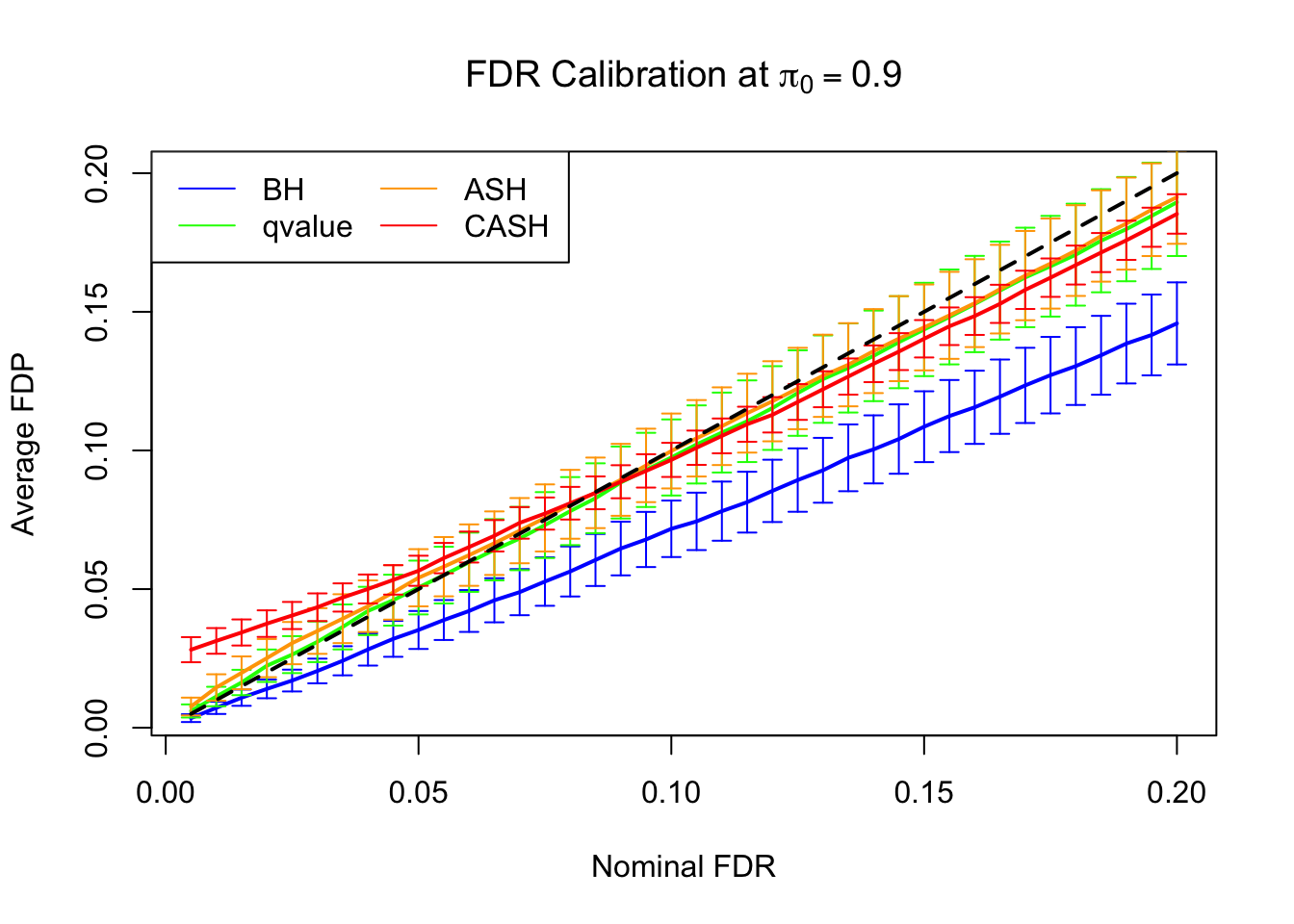
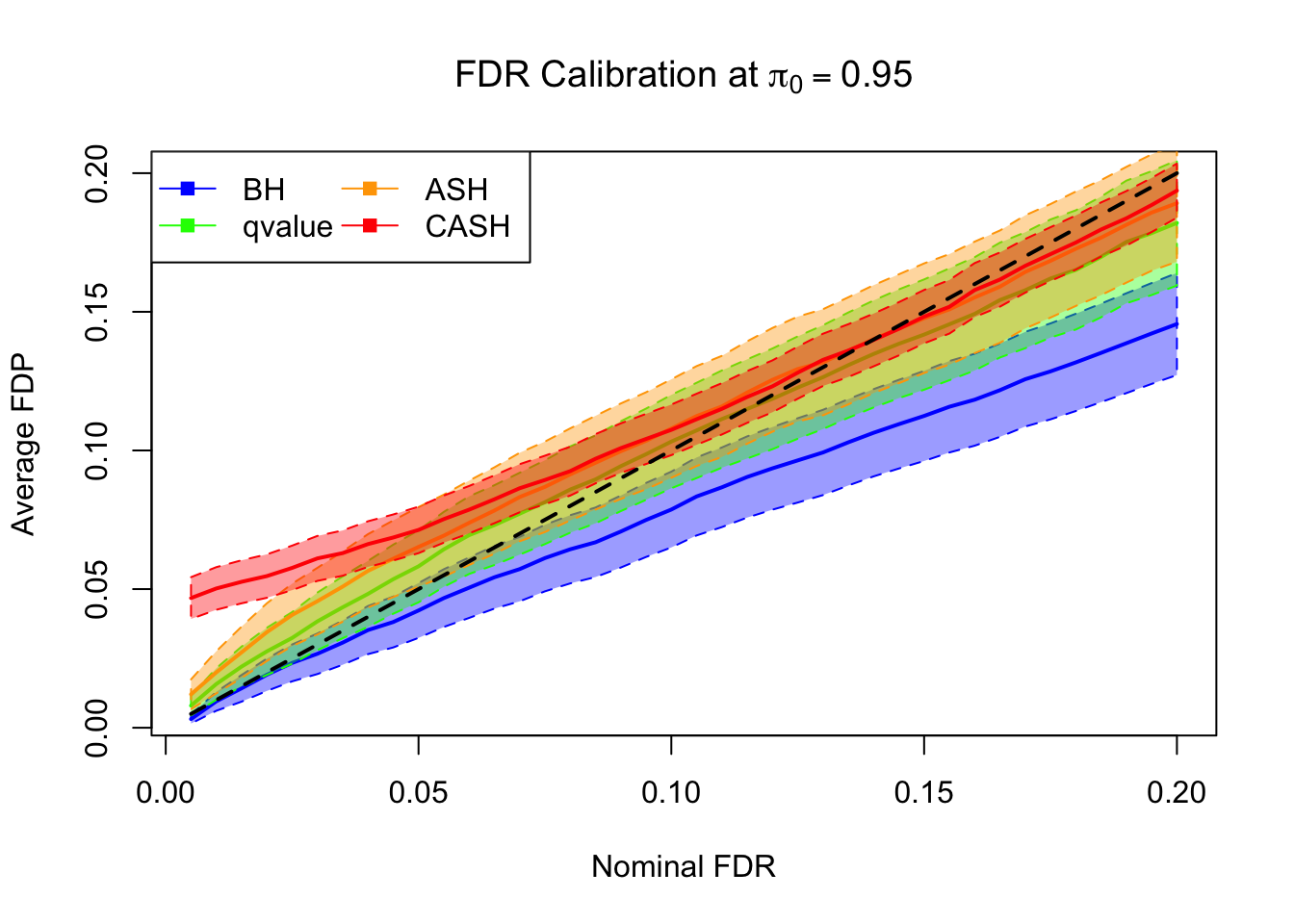
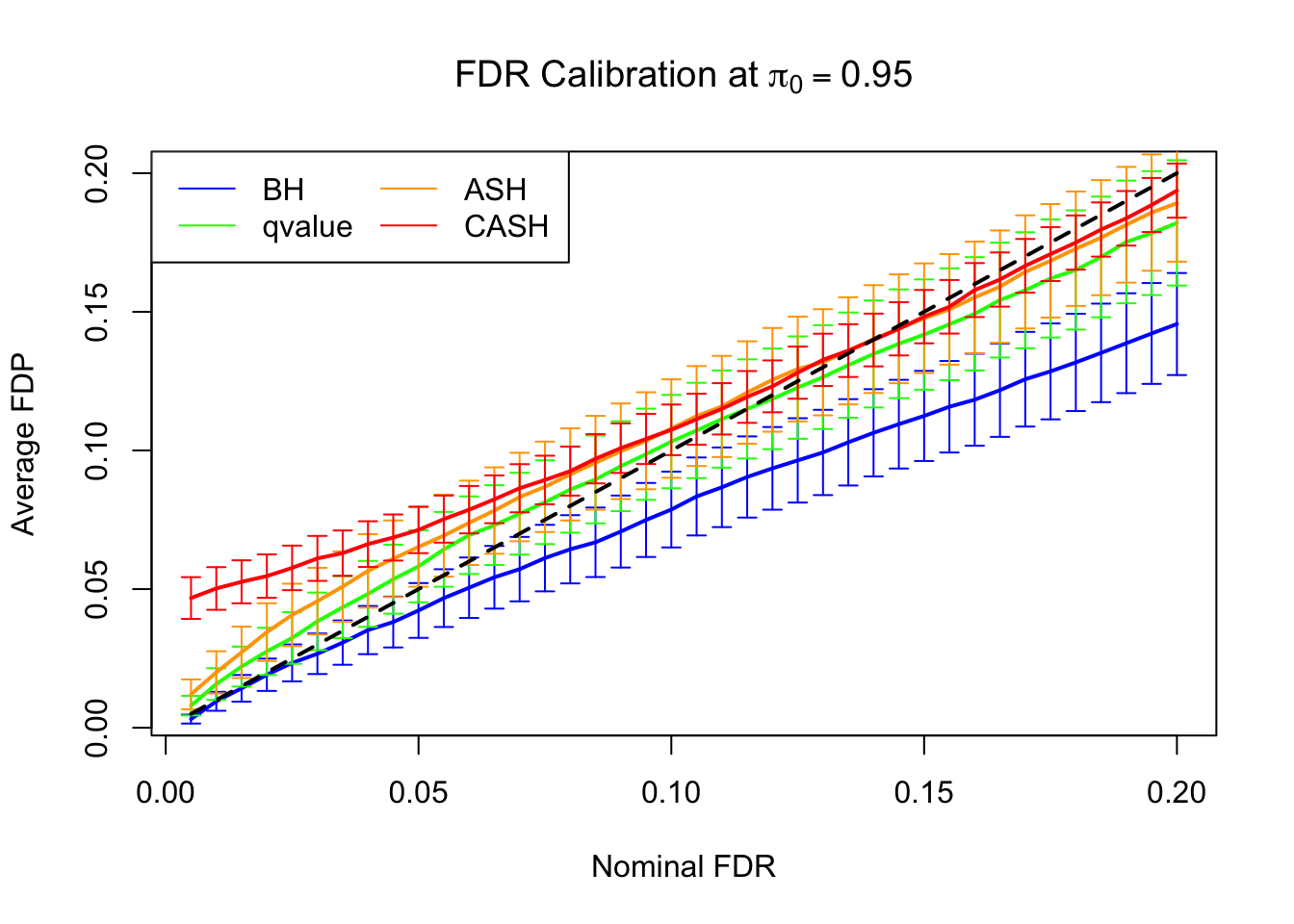
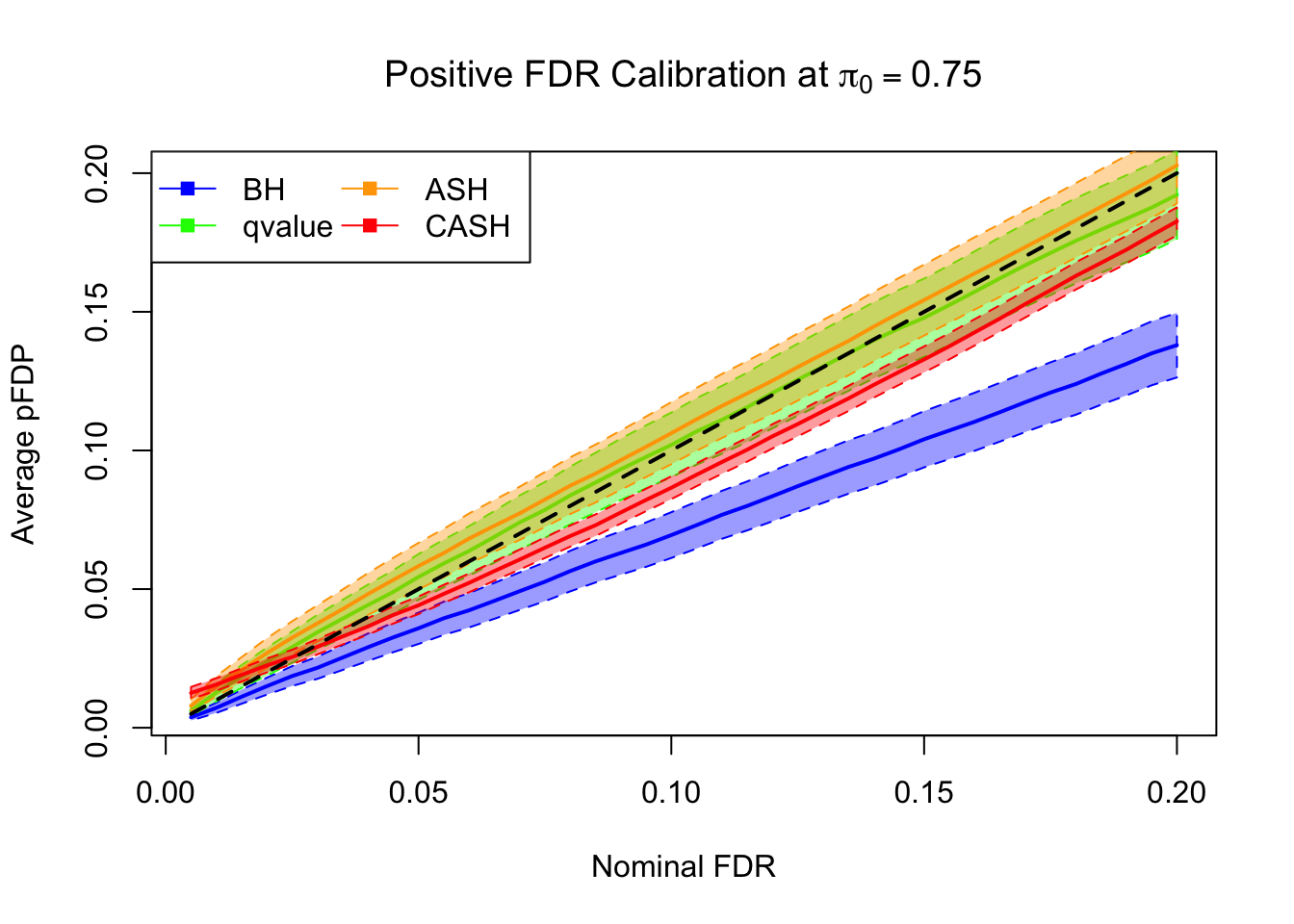
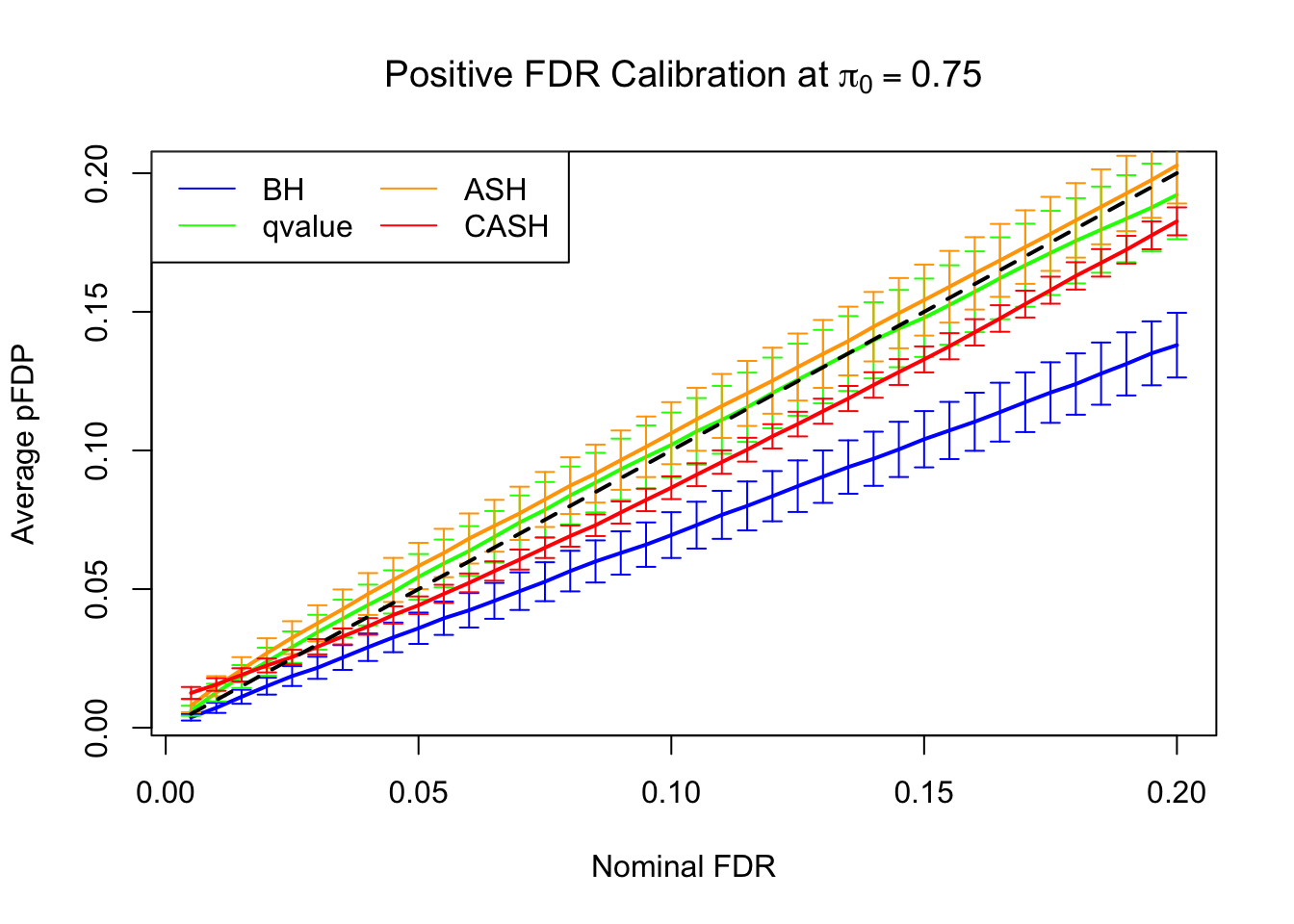
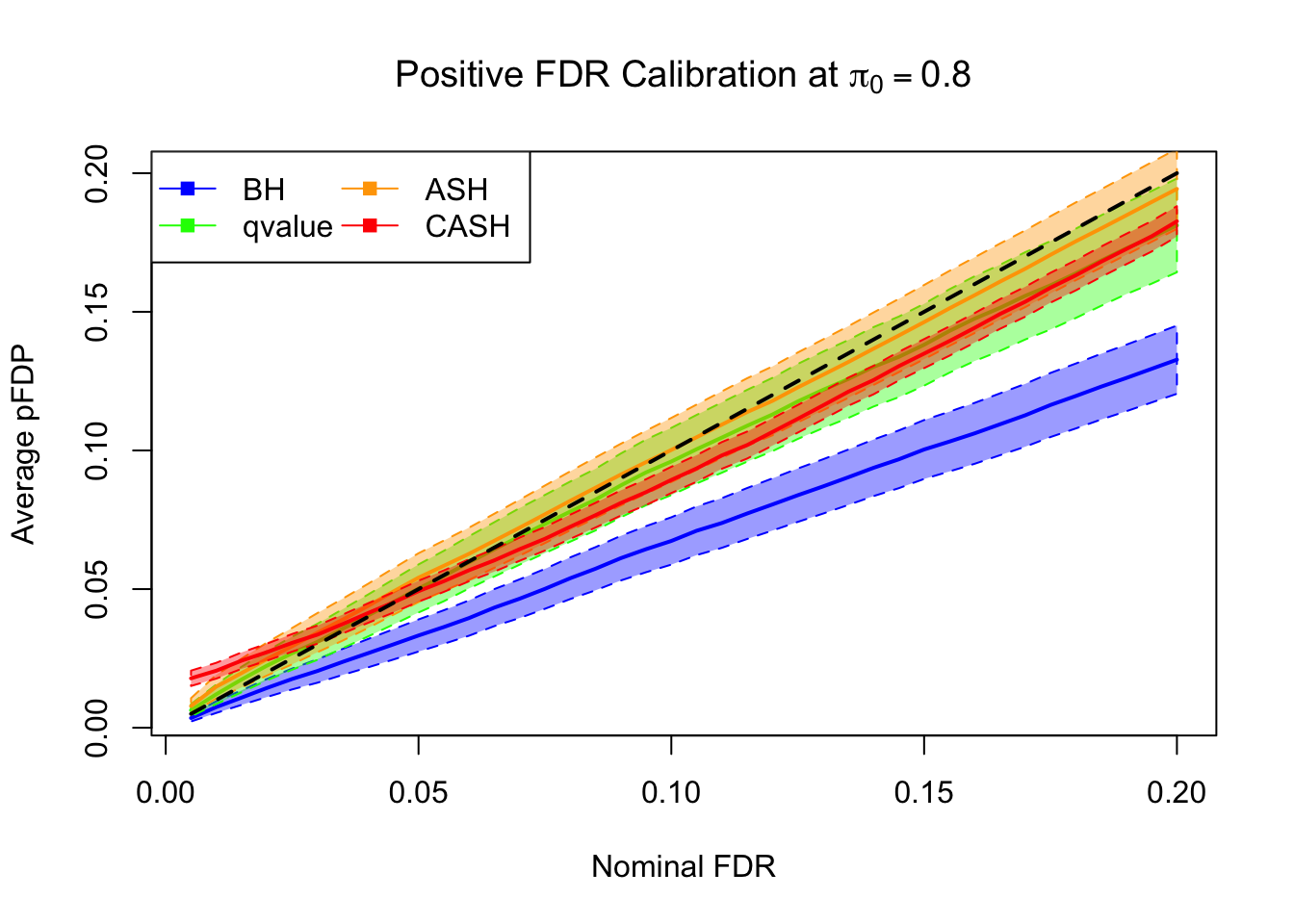
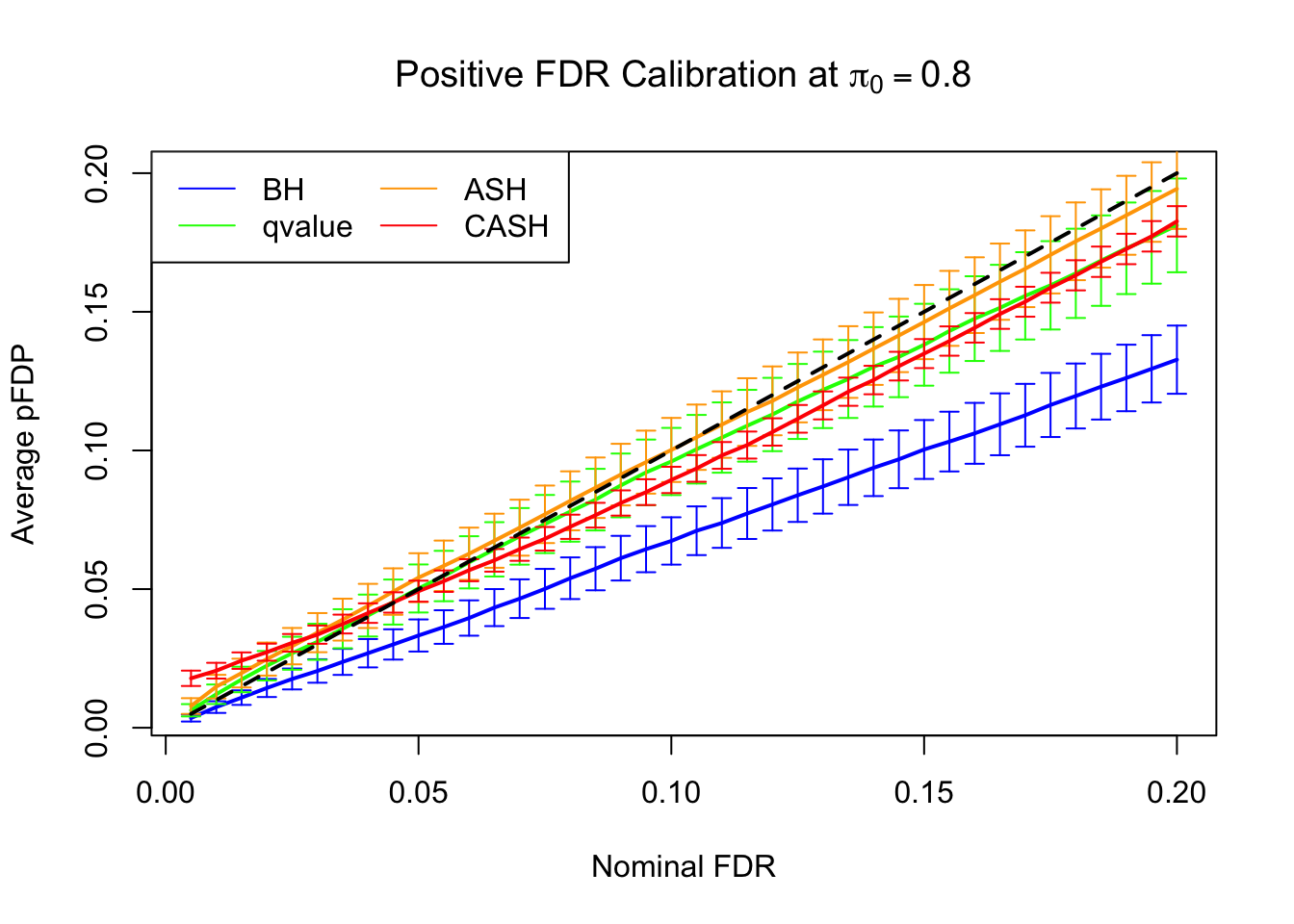
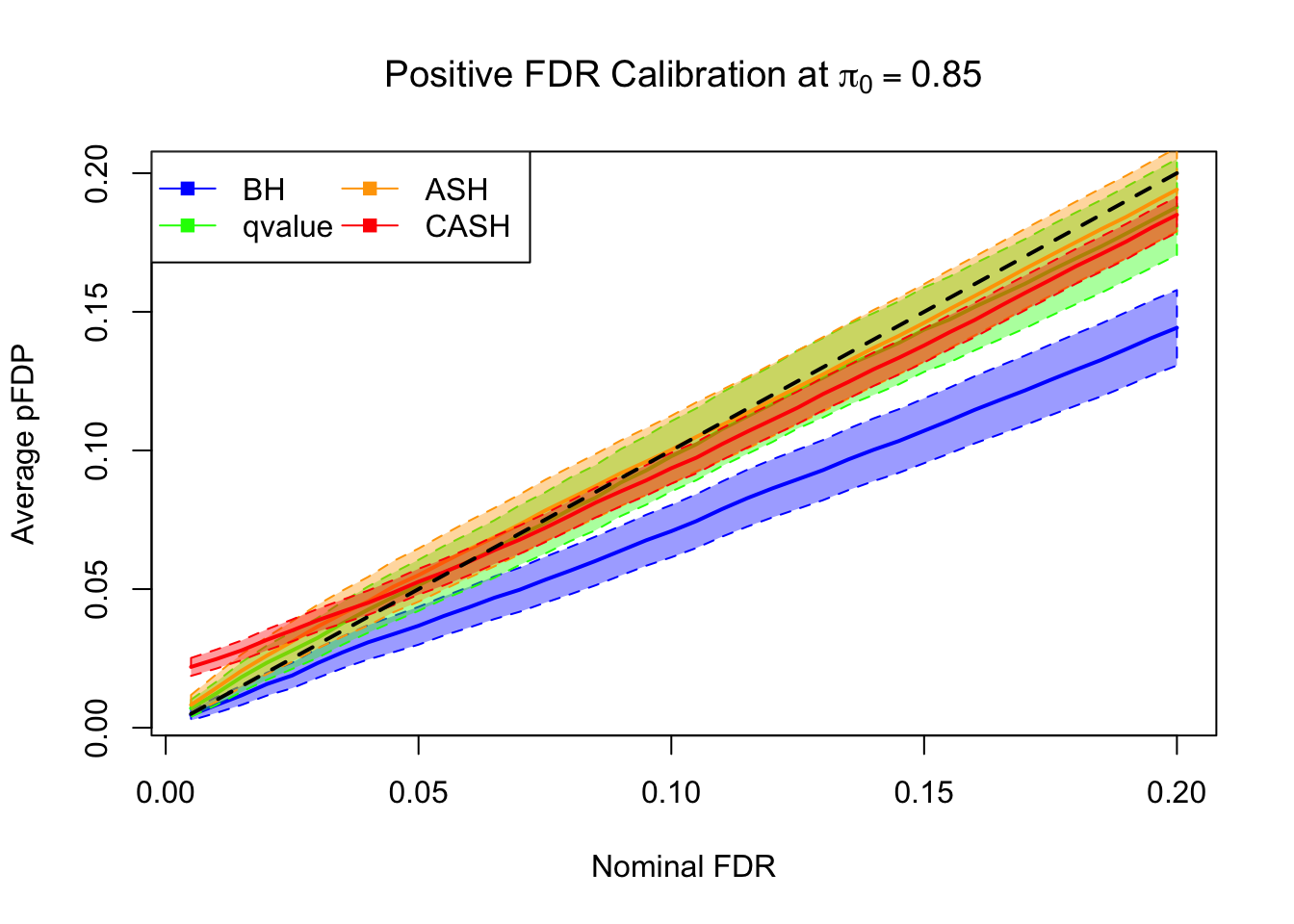
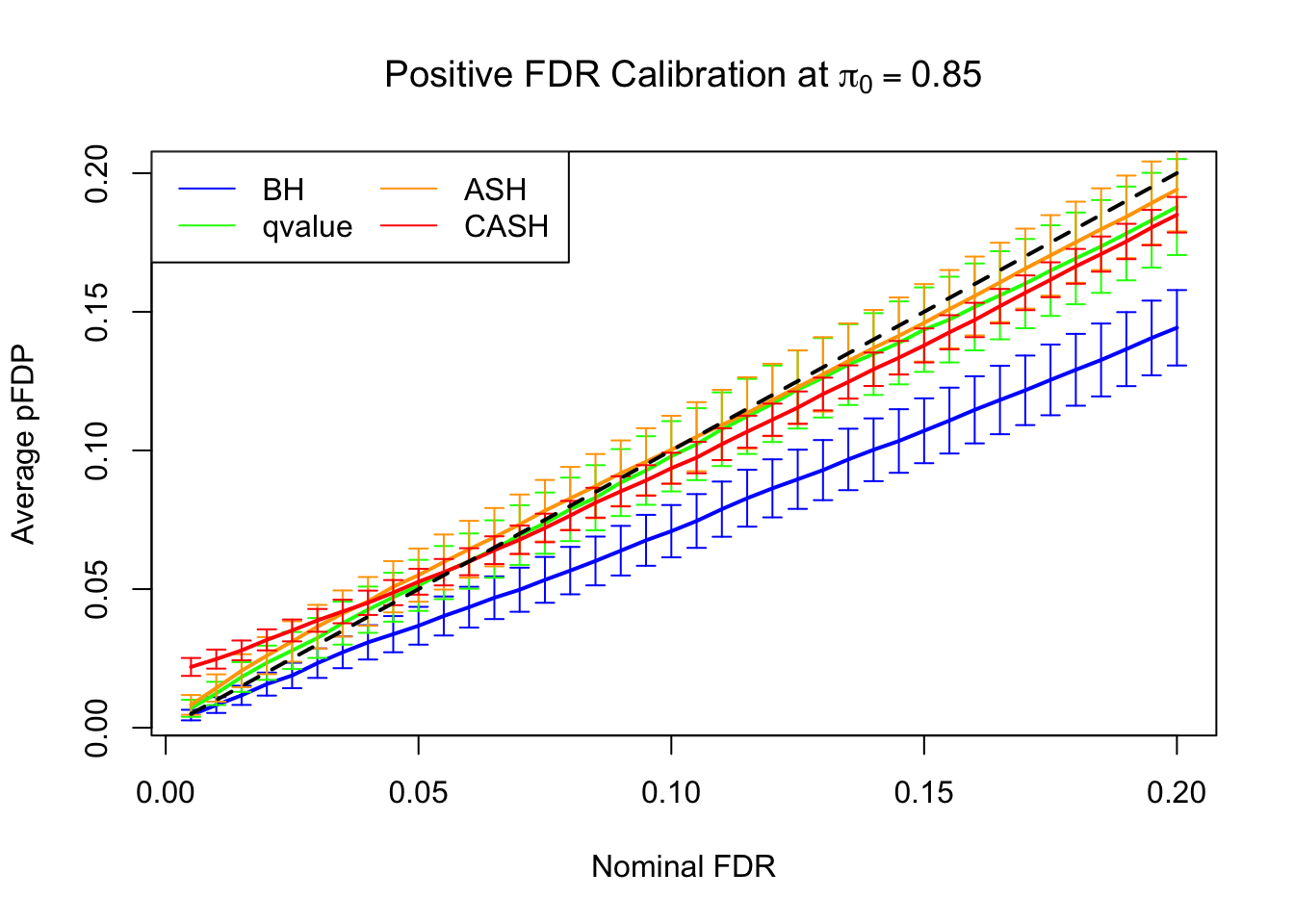
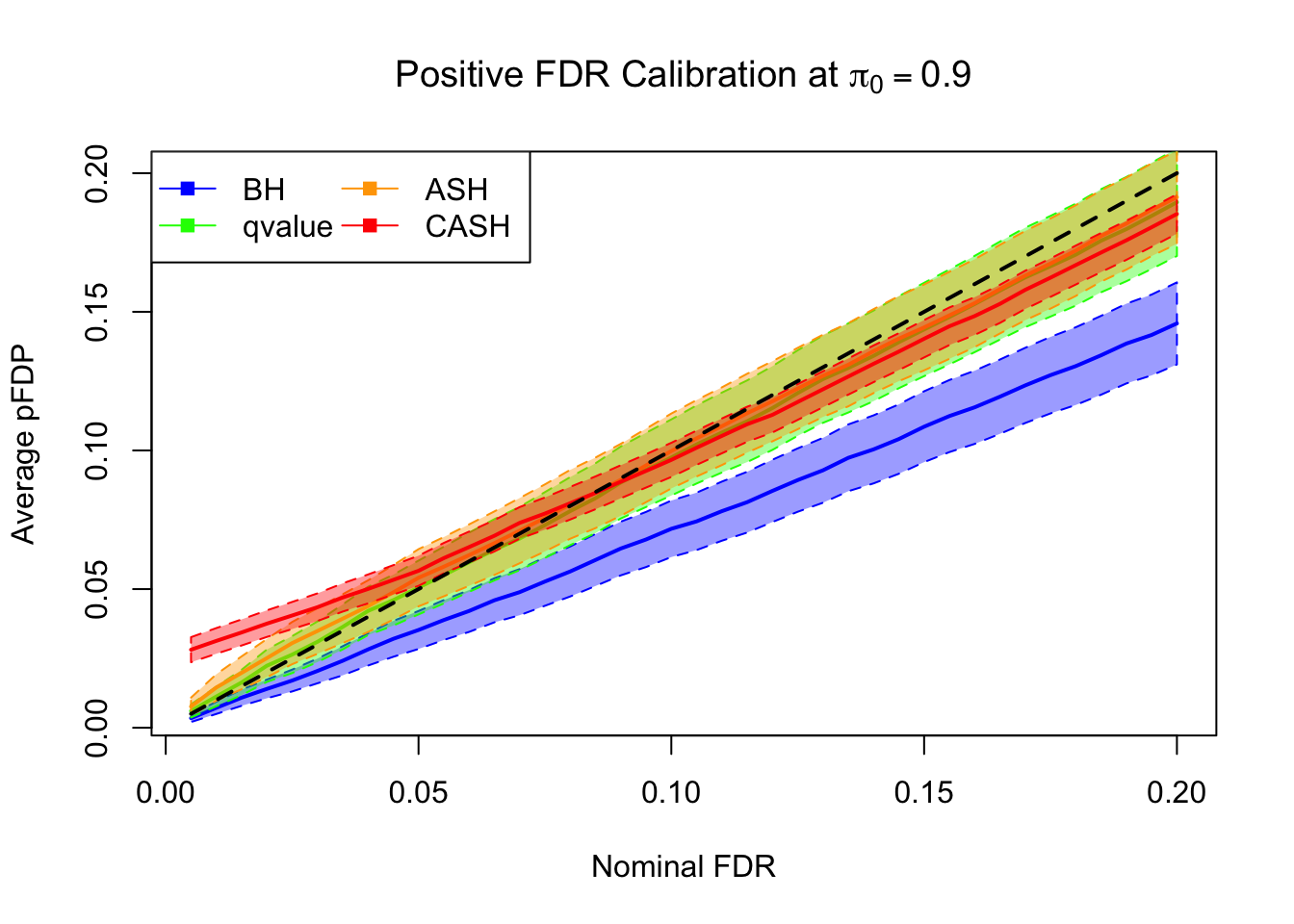
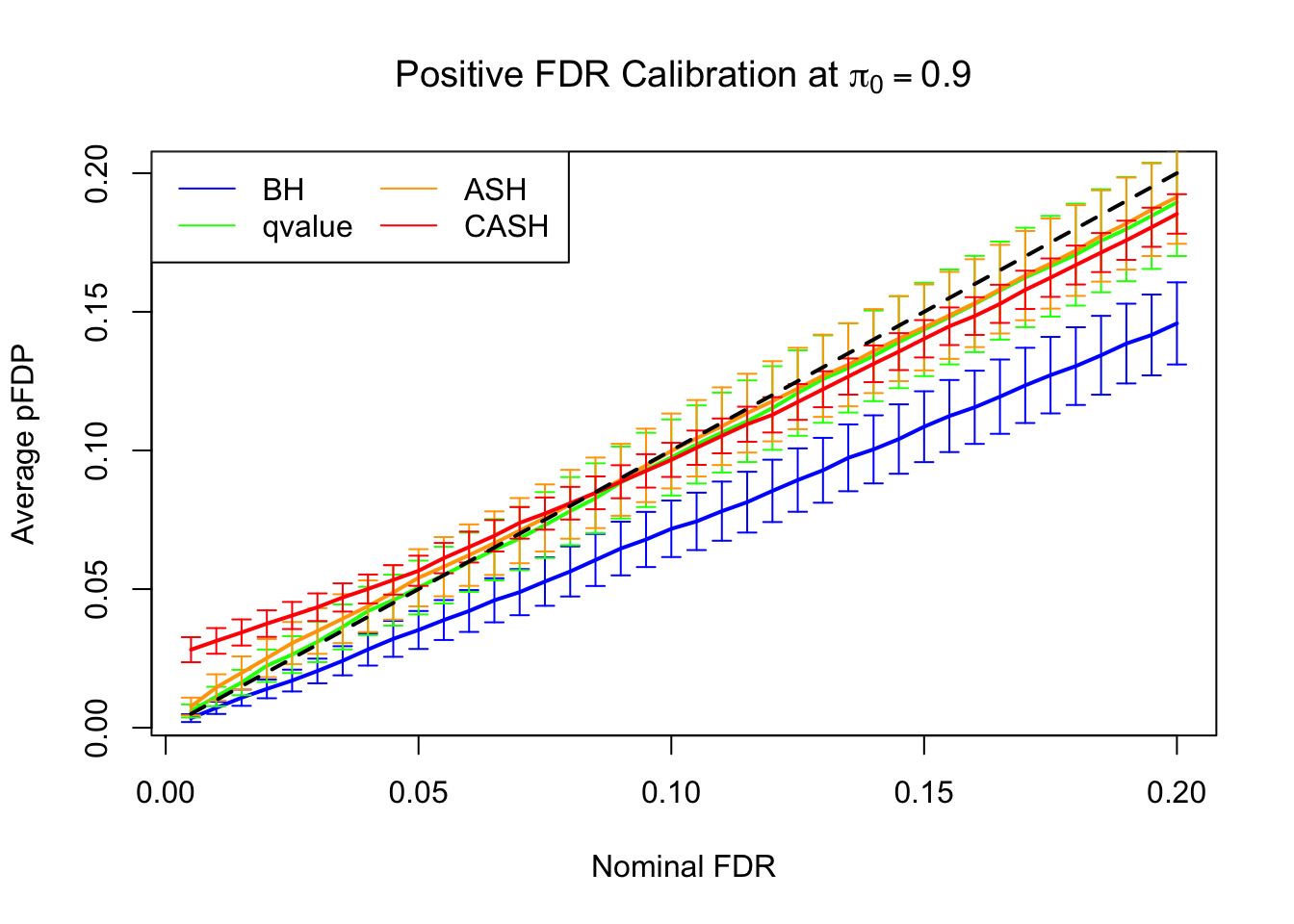
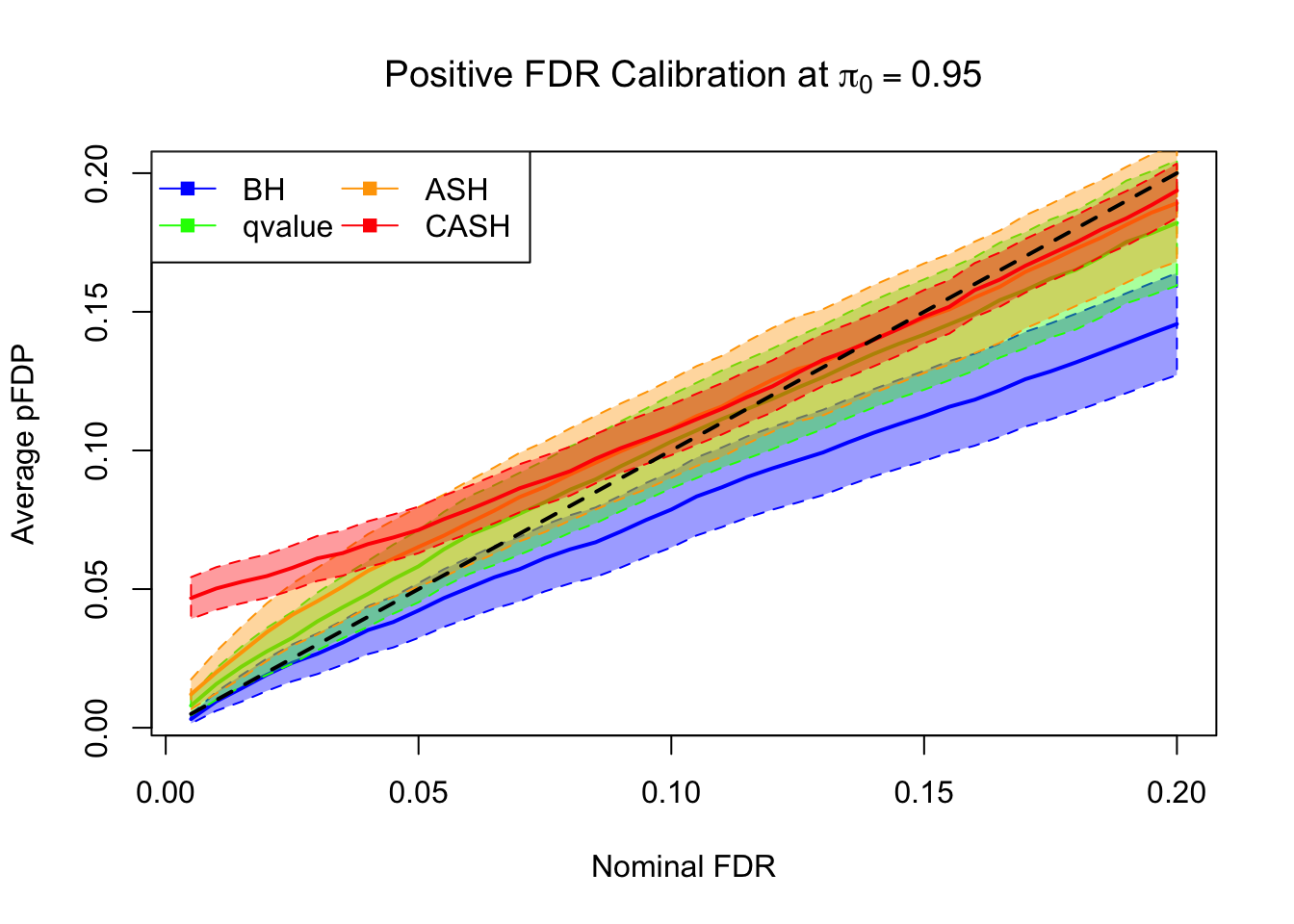
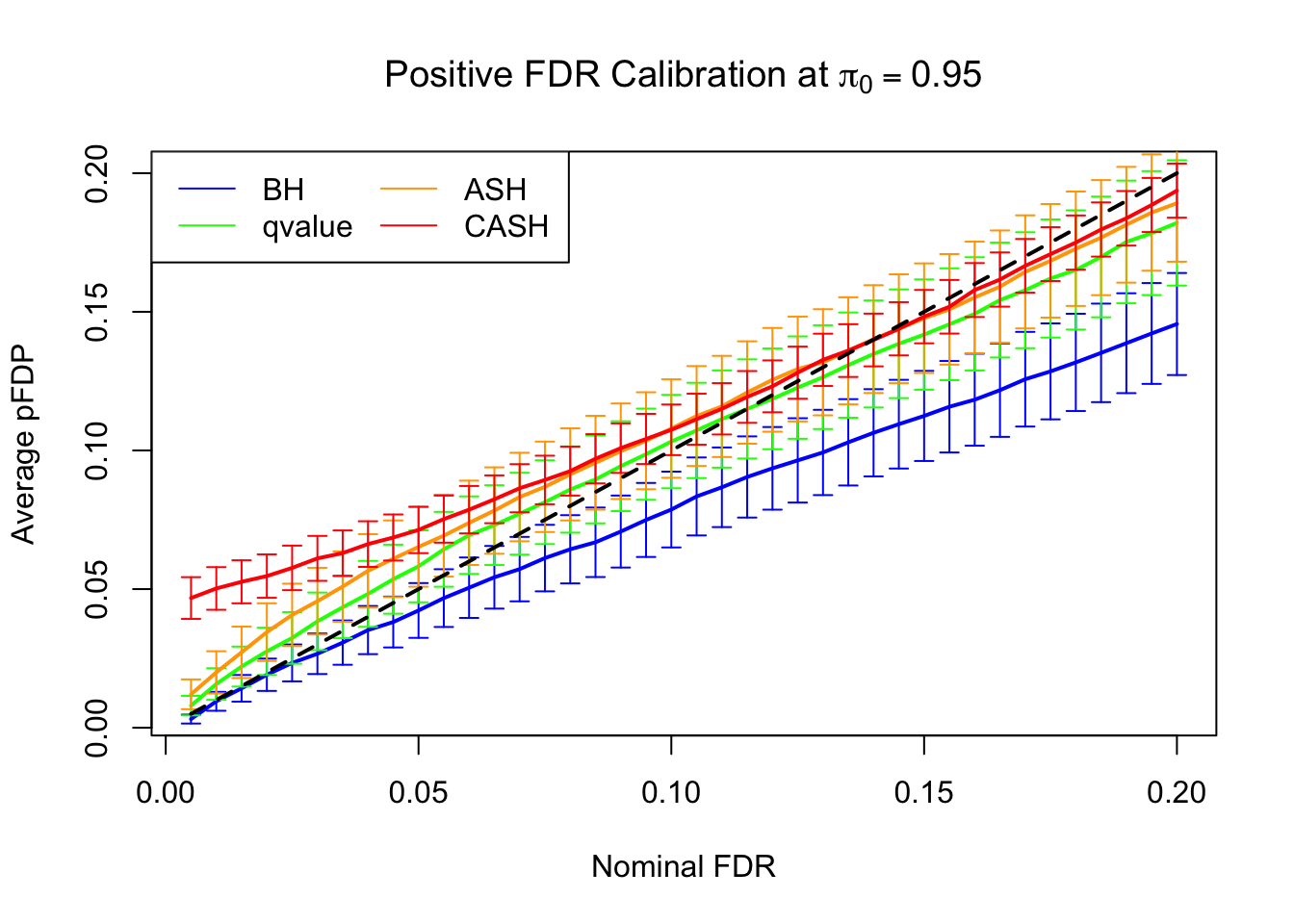
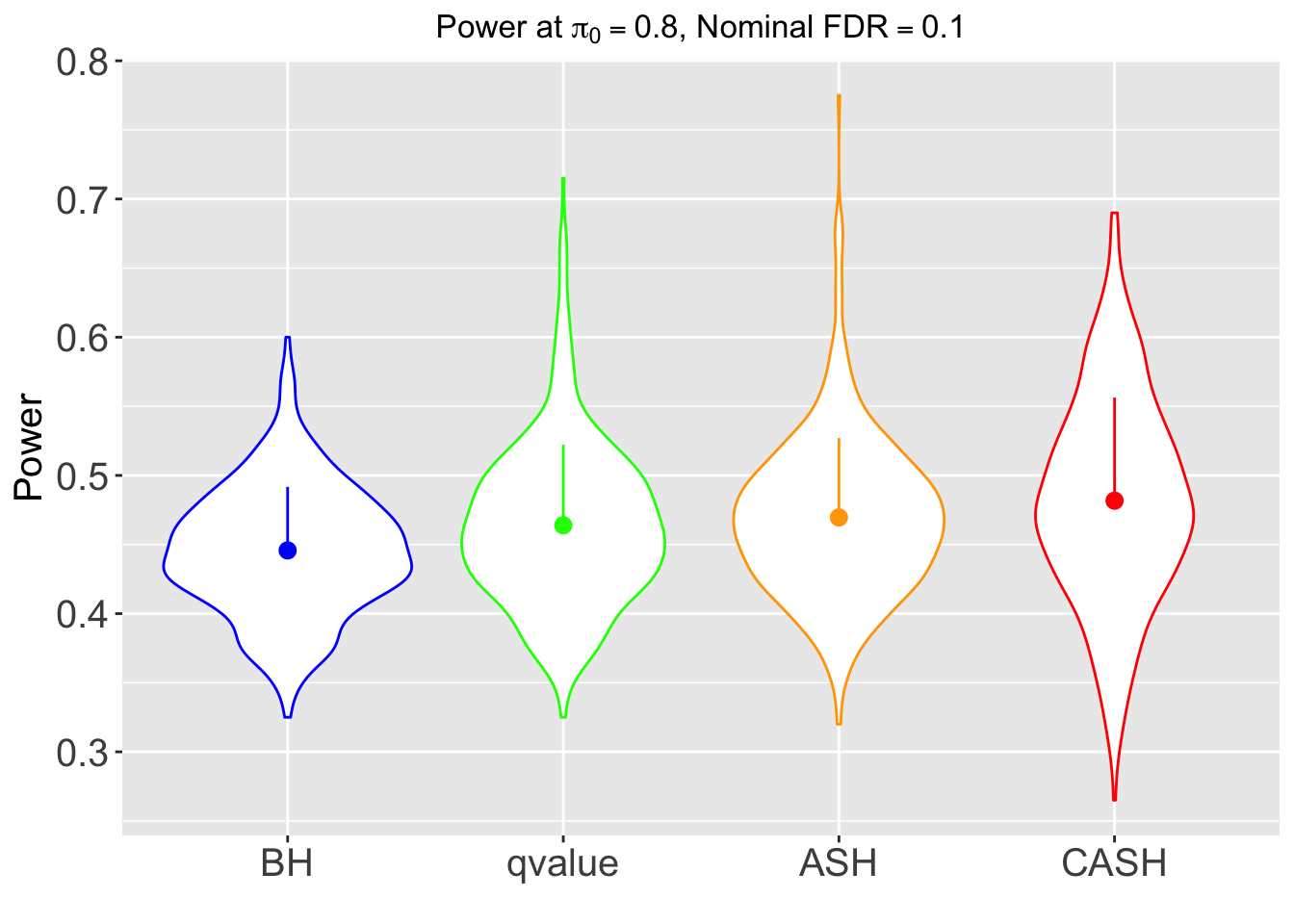
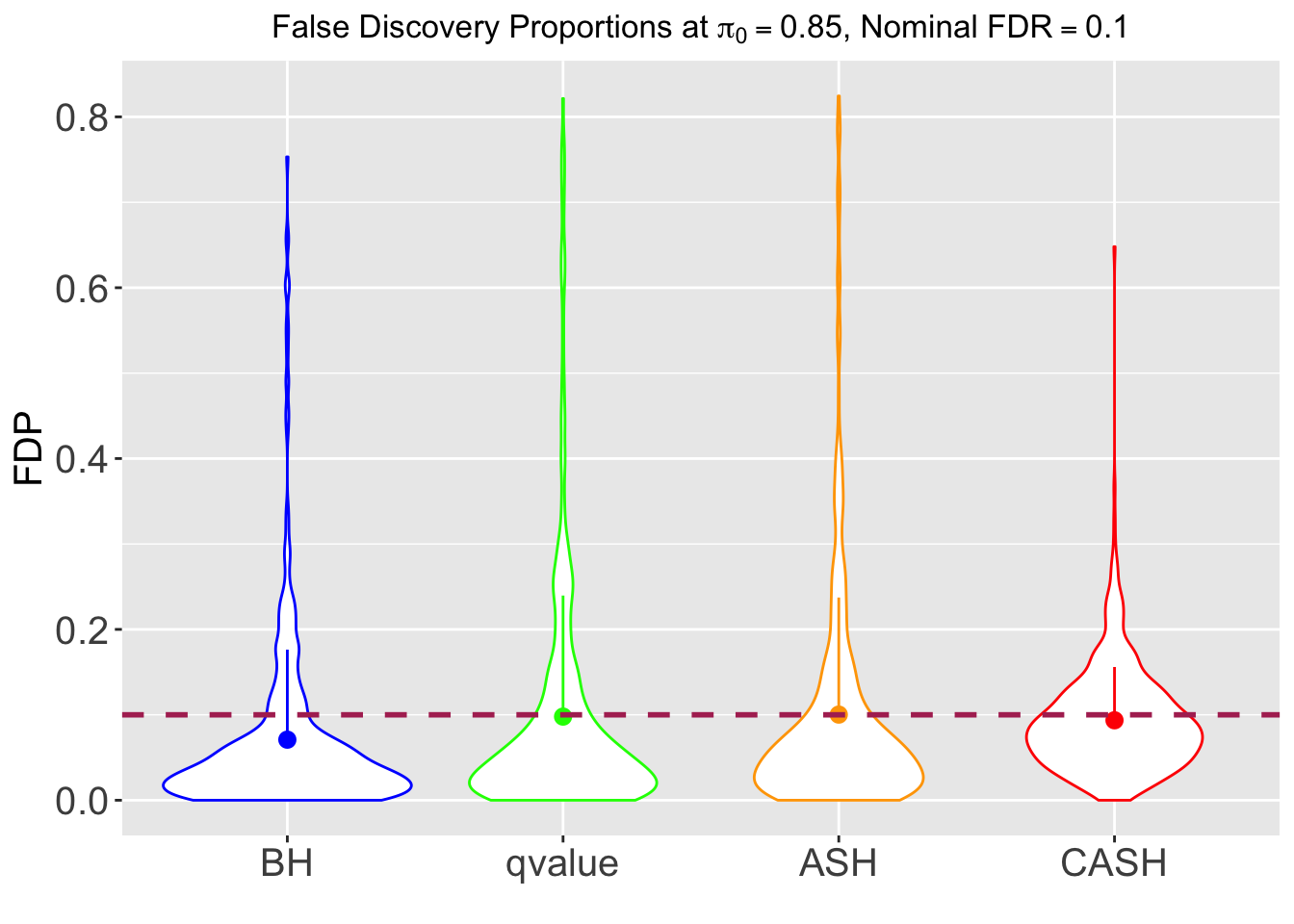
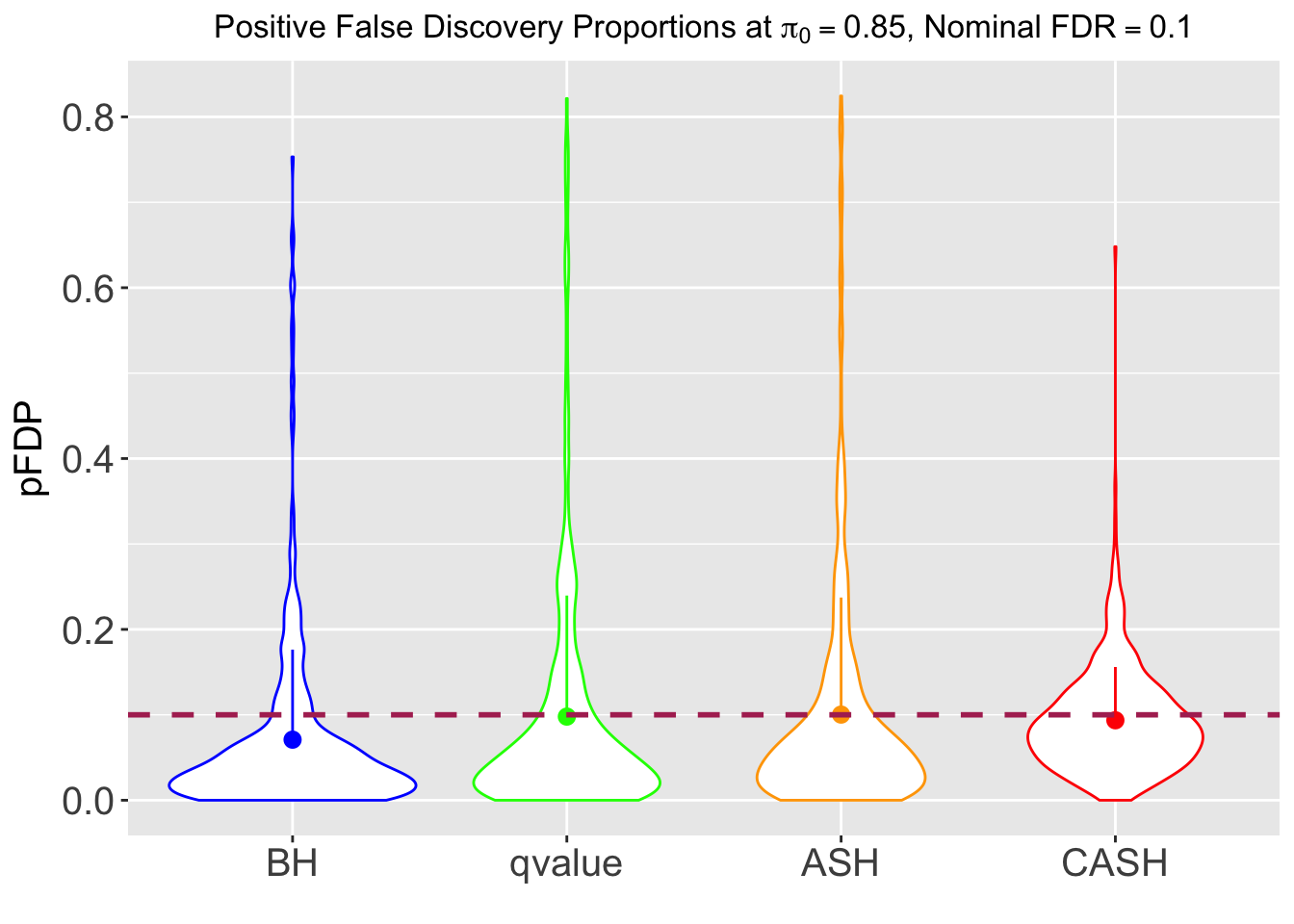
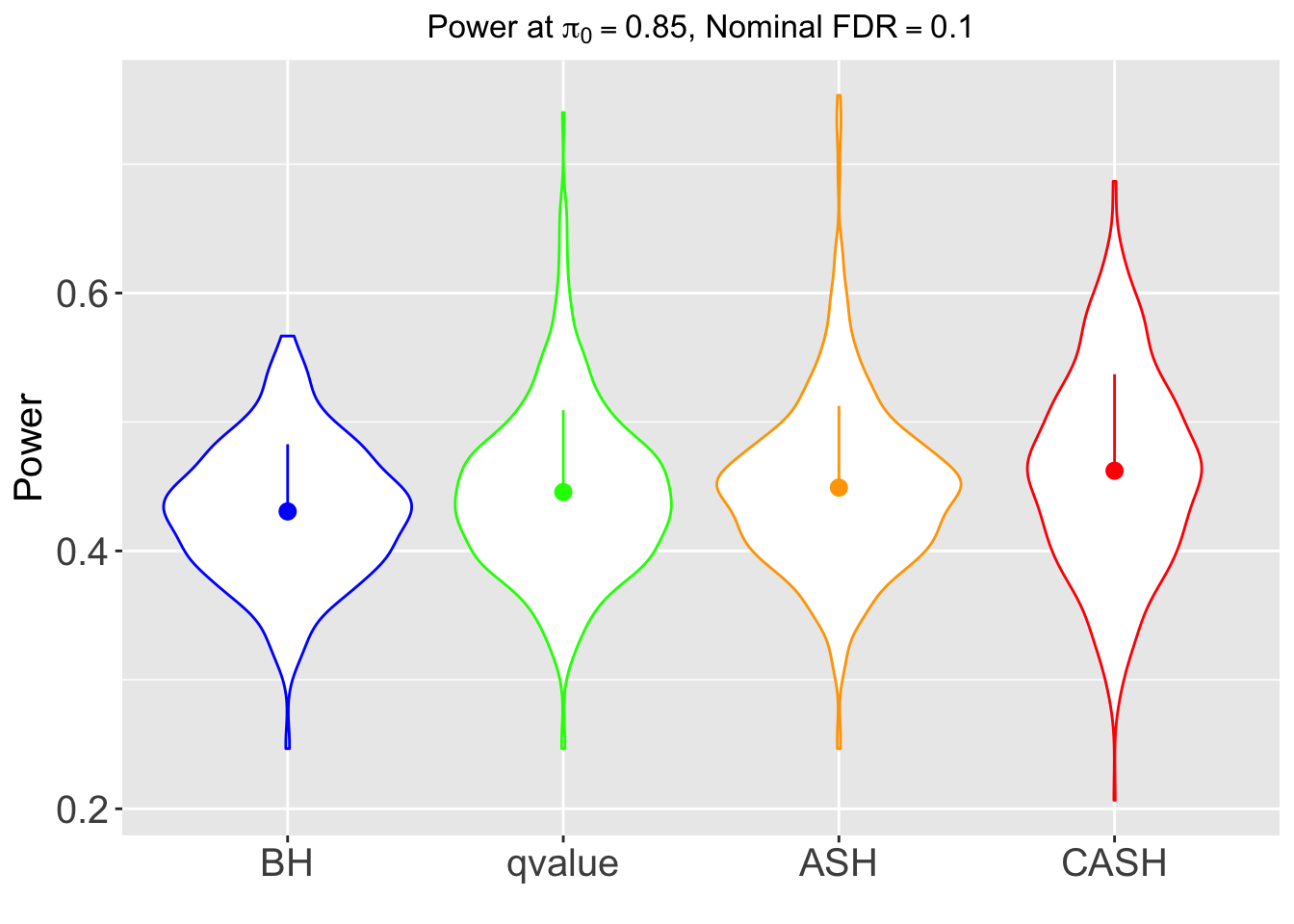
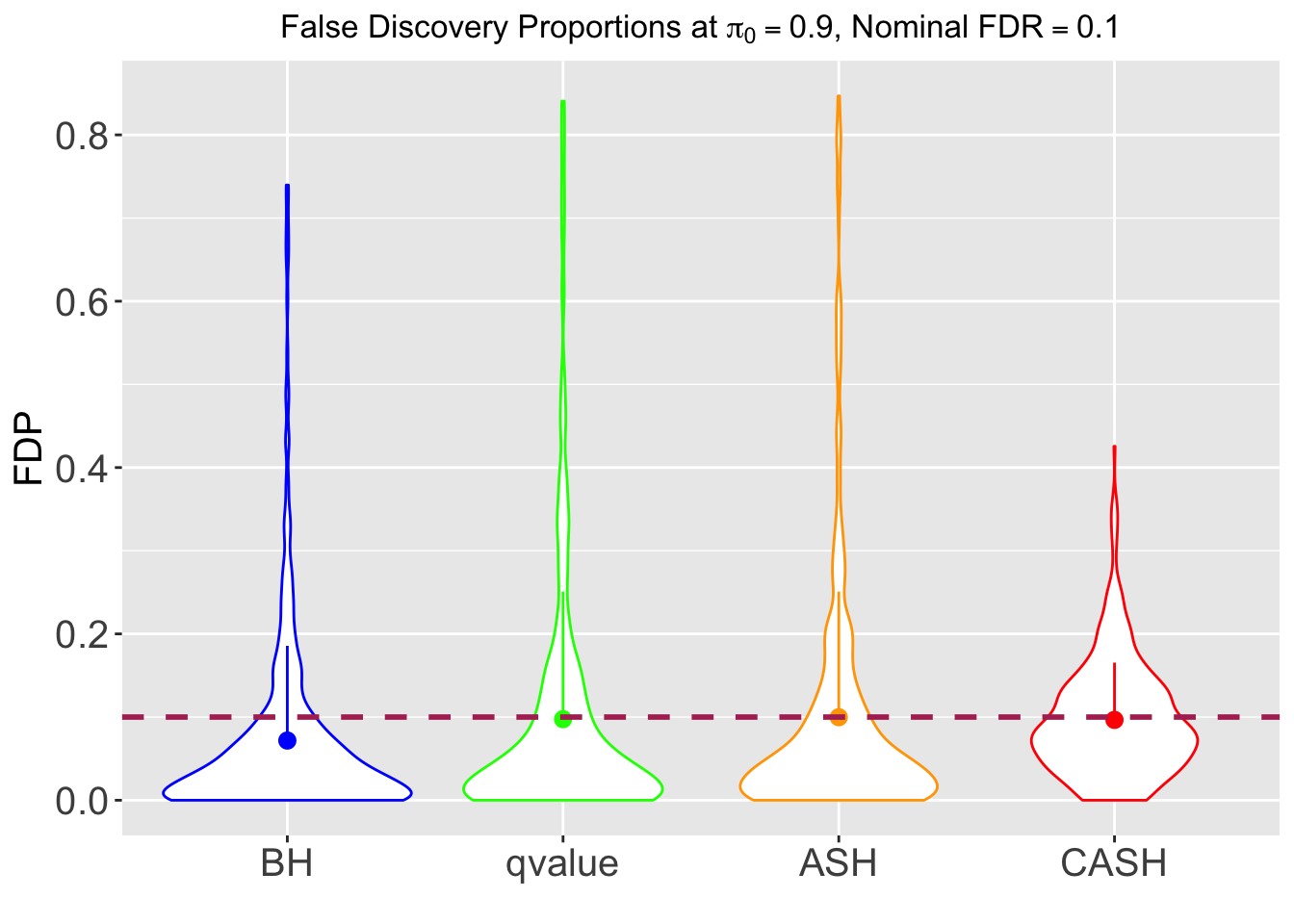
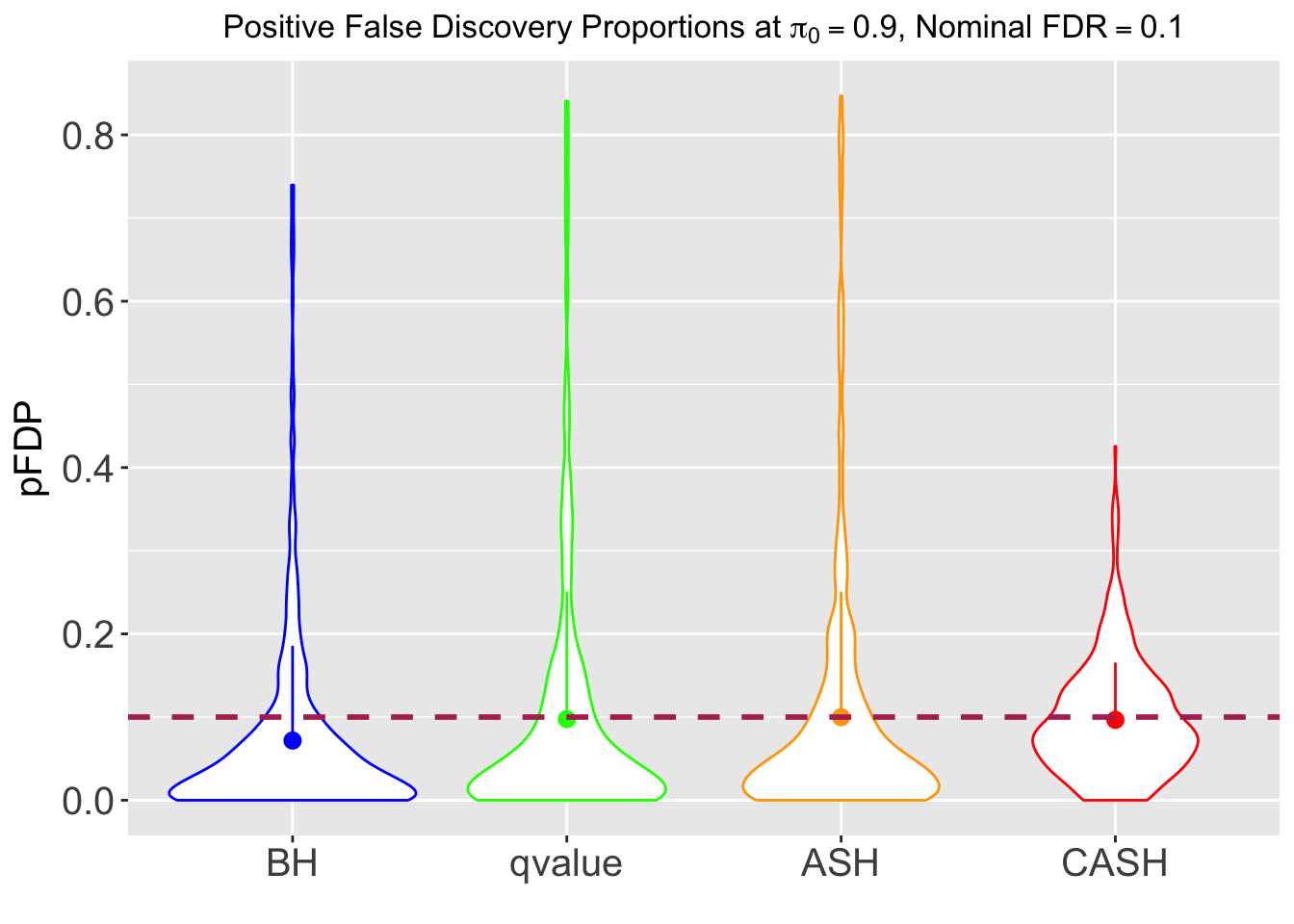
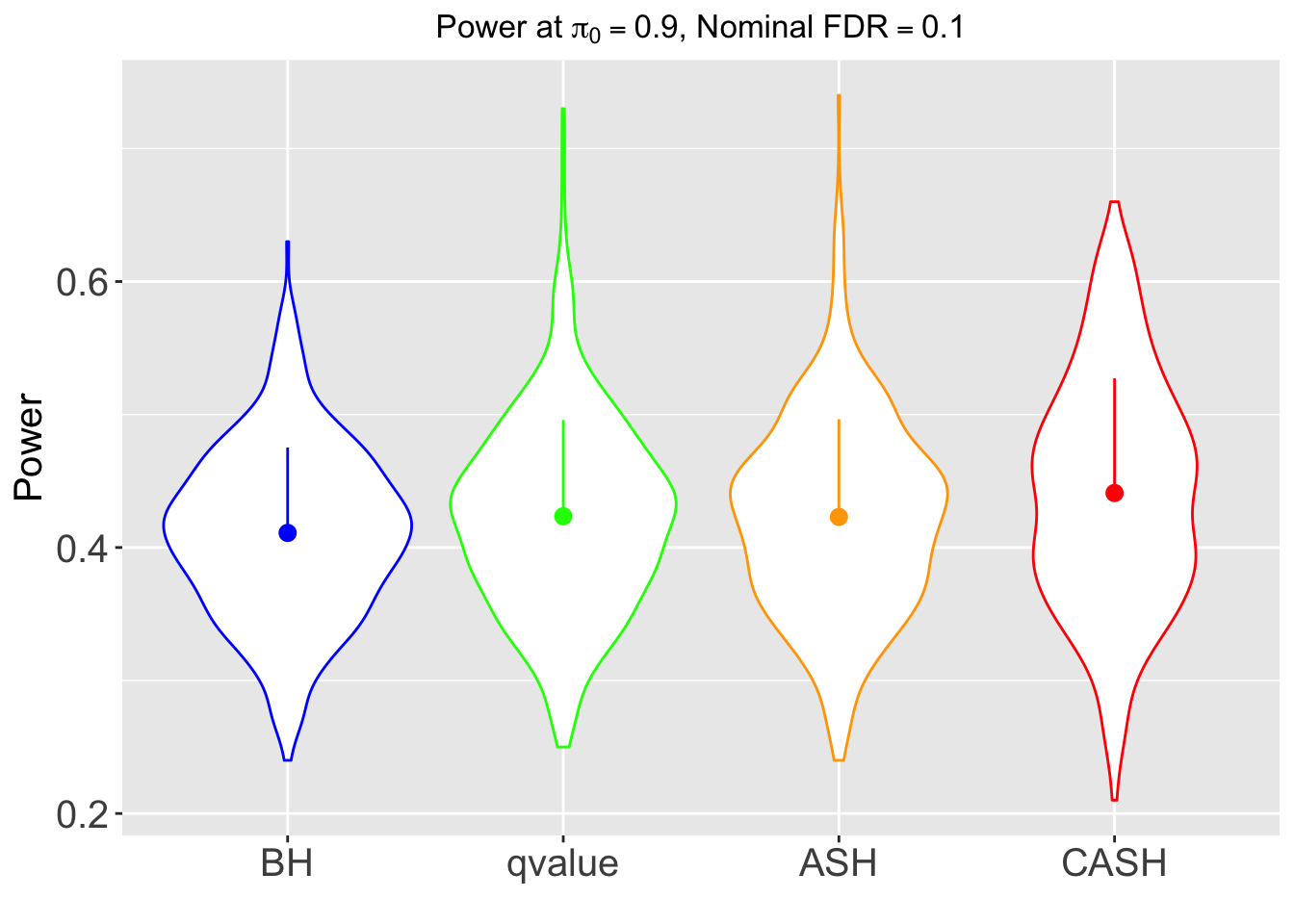
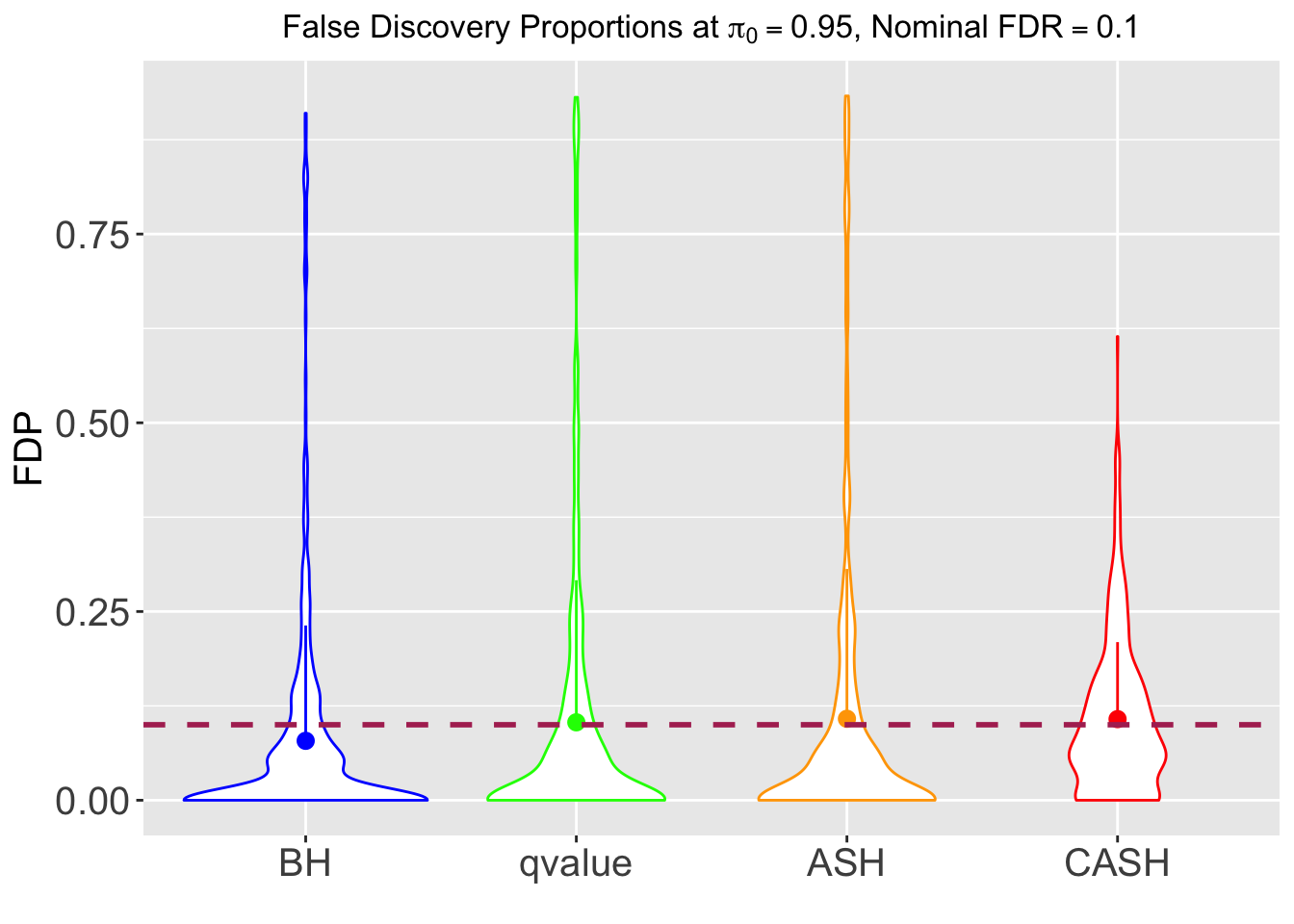
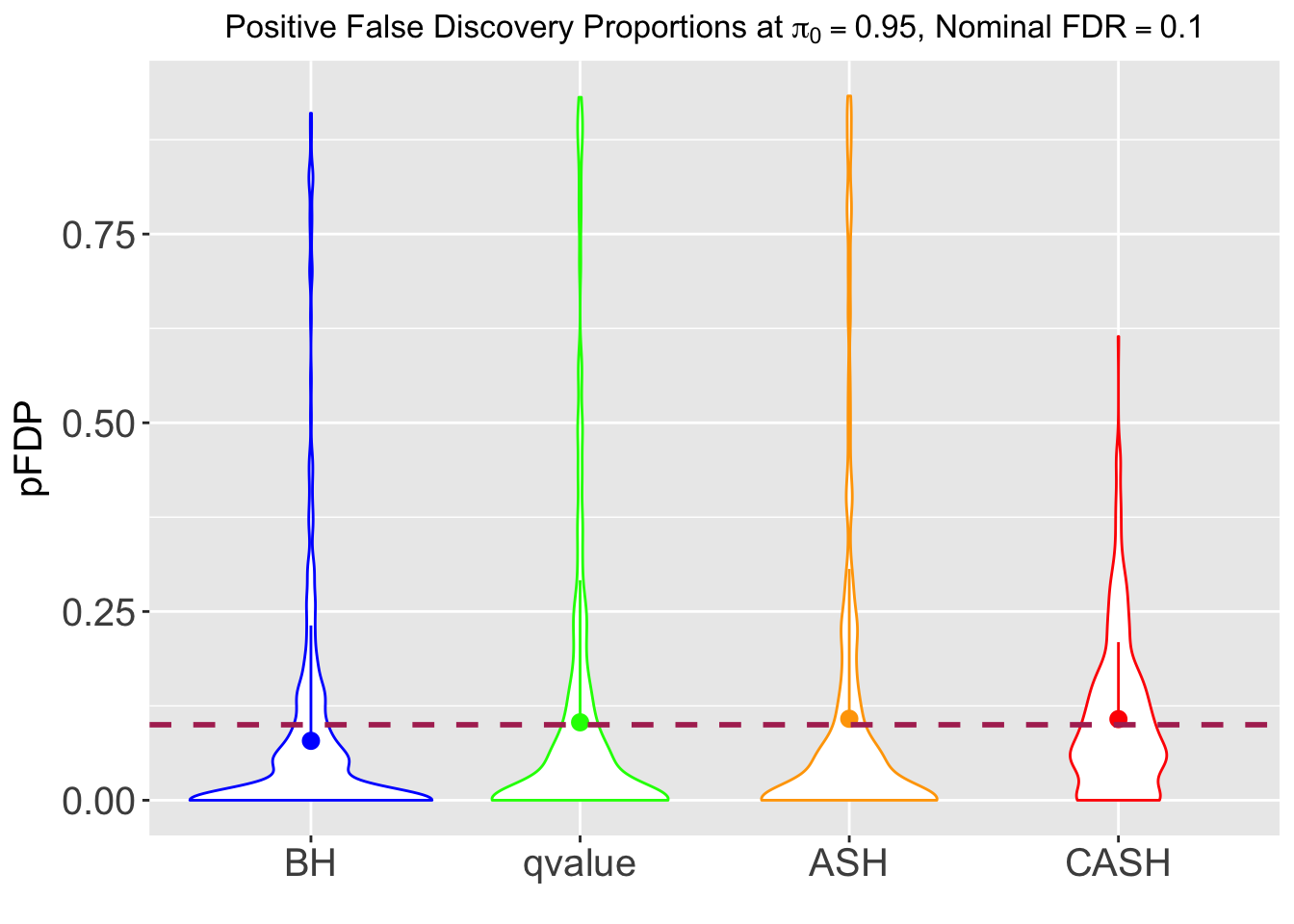
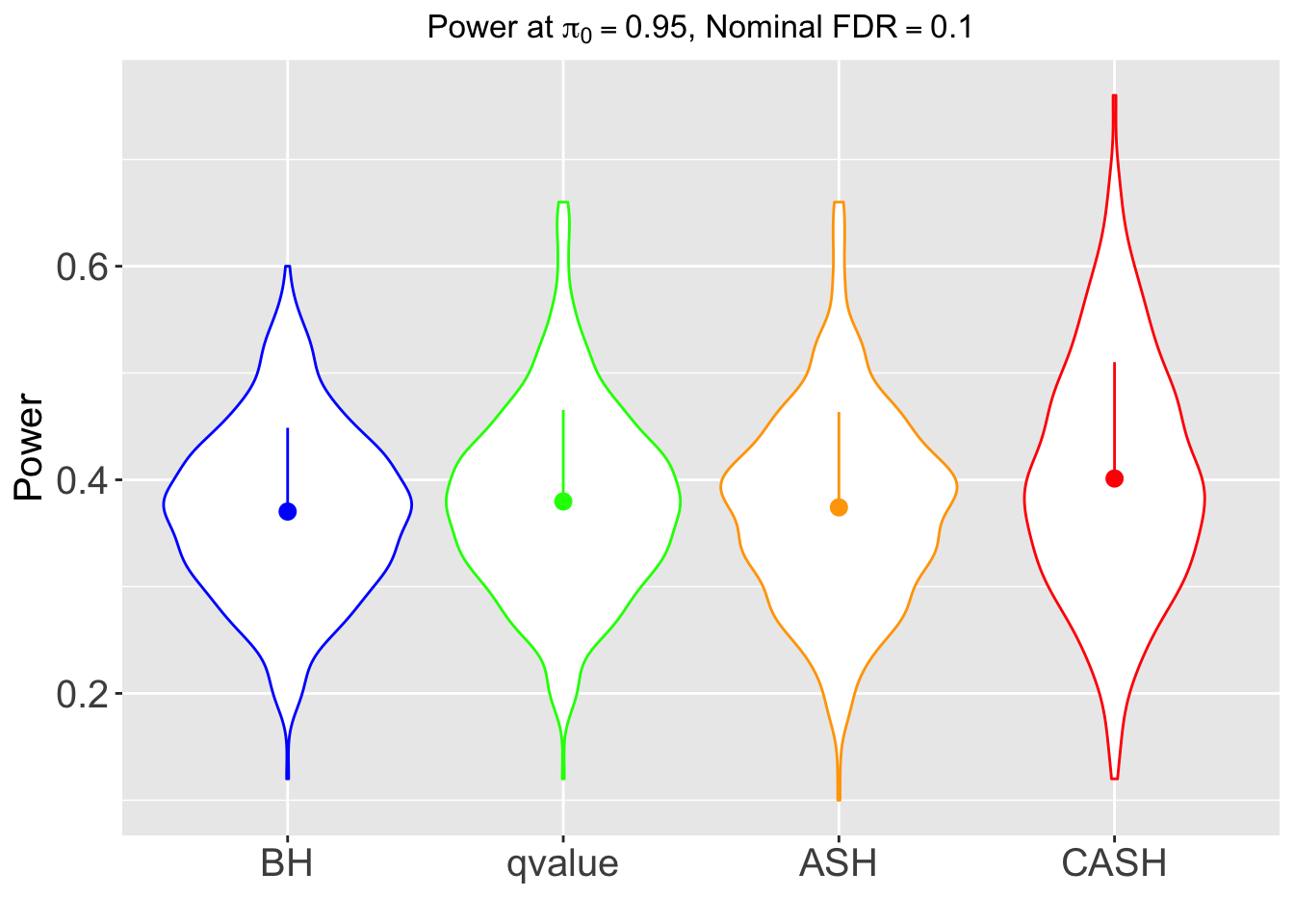
FDR, pFDR, Power at Nominal FDR = 0.1
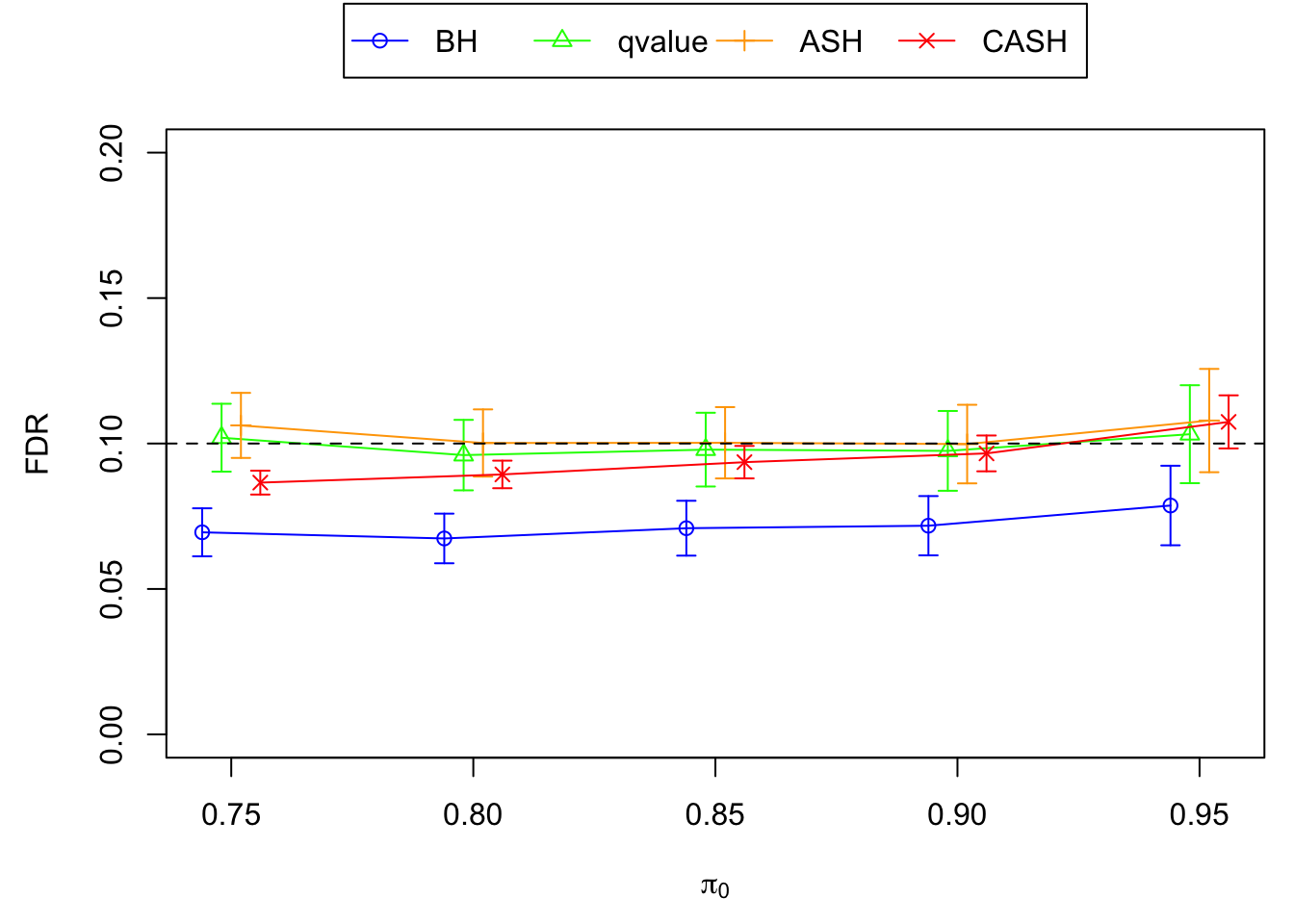
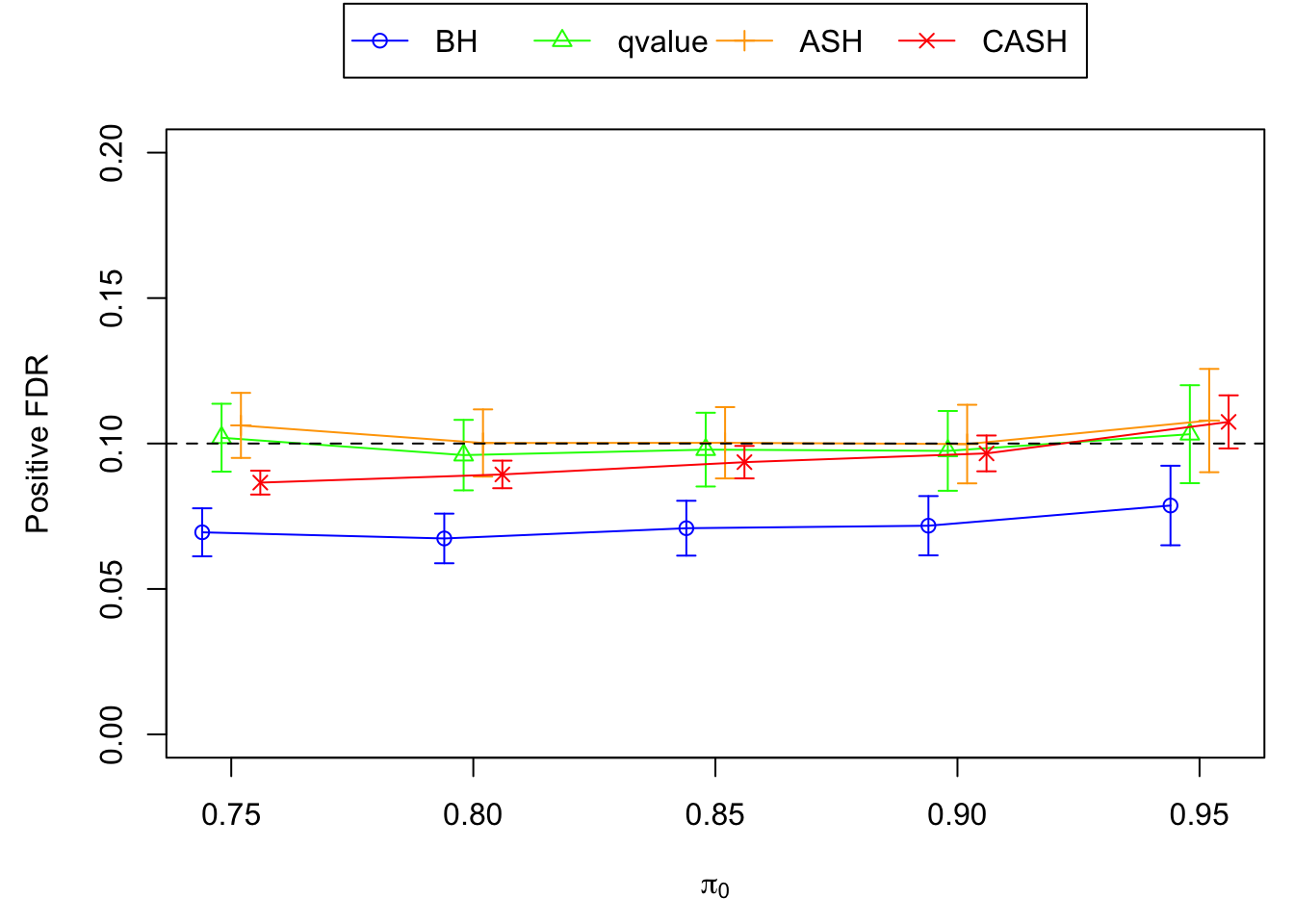
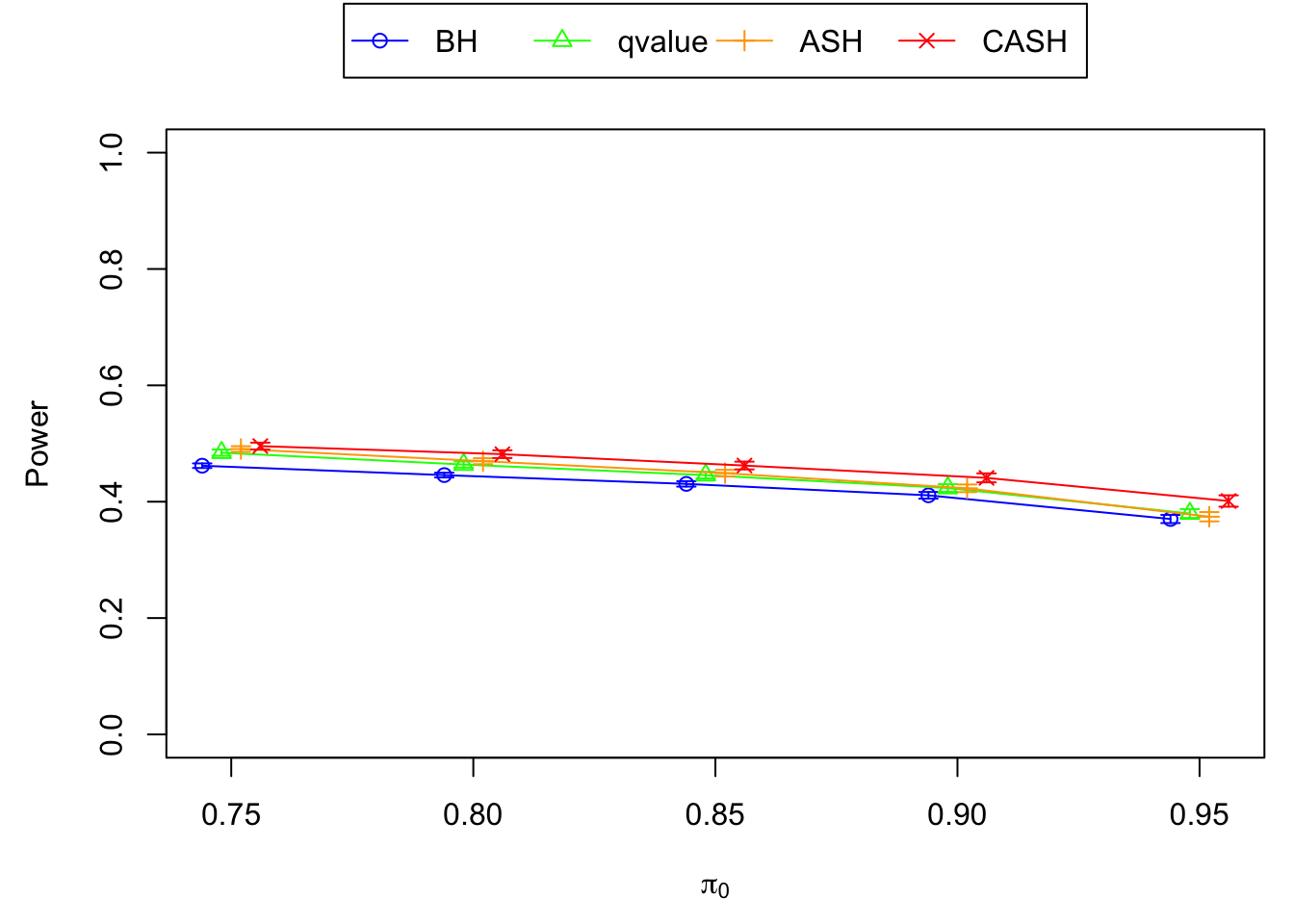
FDR, Power at each sparsity level at Nominal FDR = 0.1
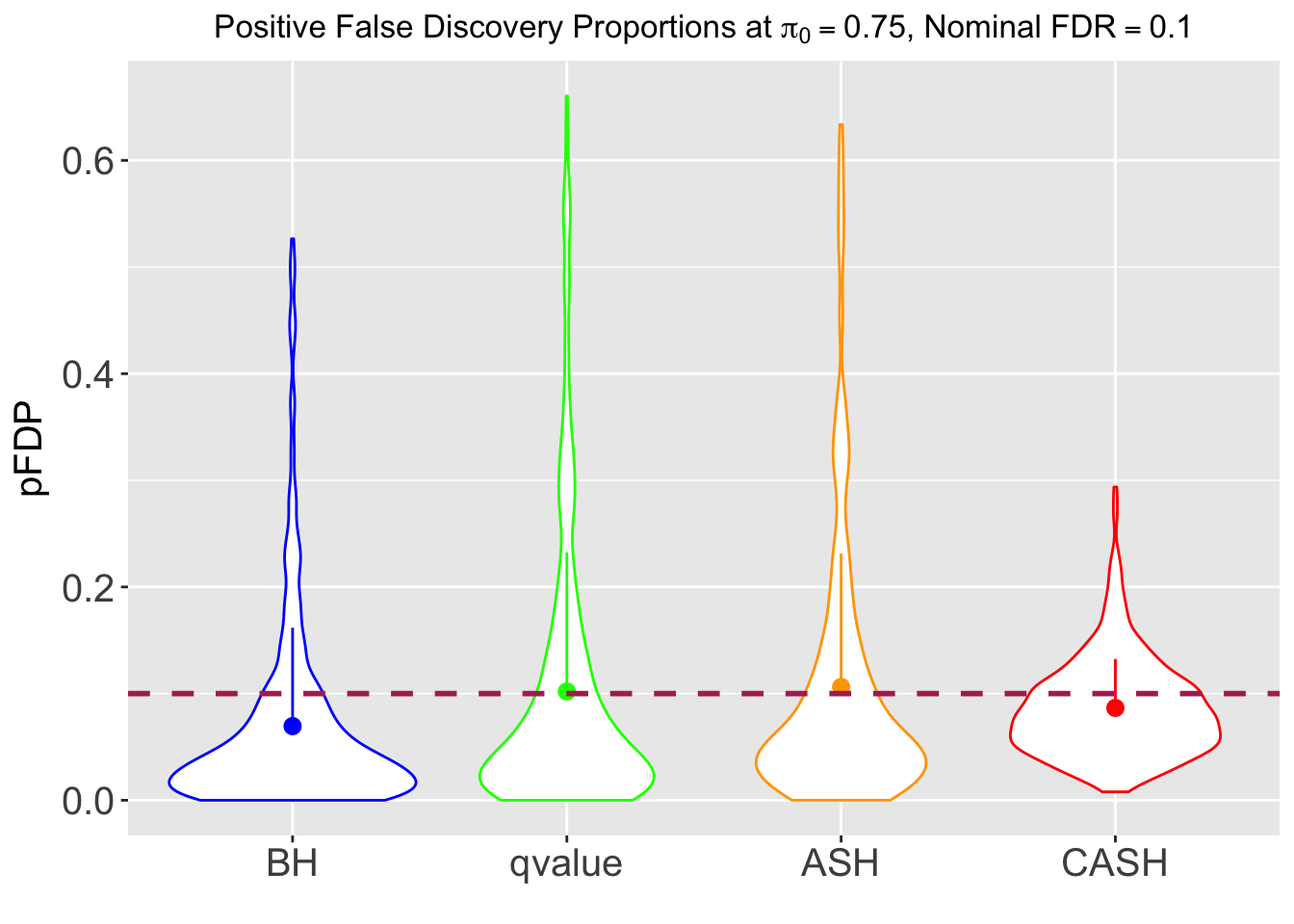
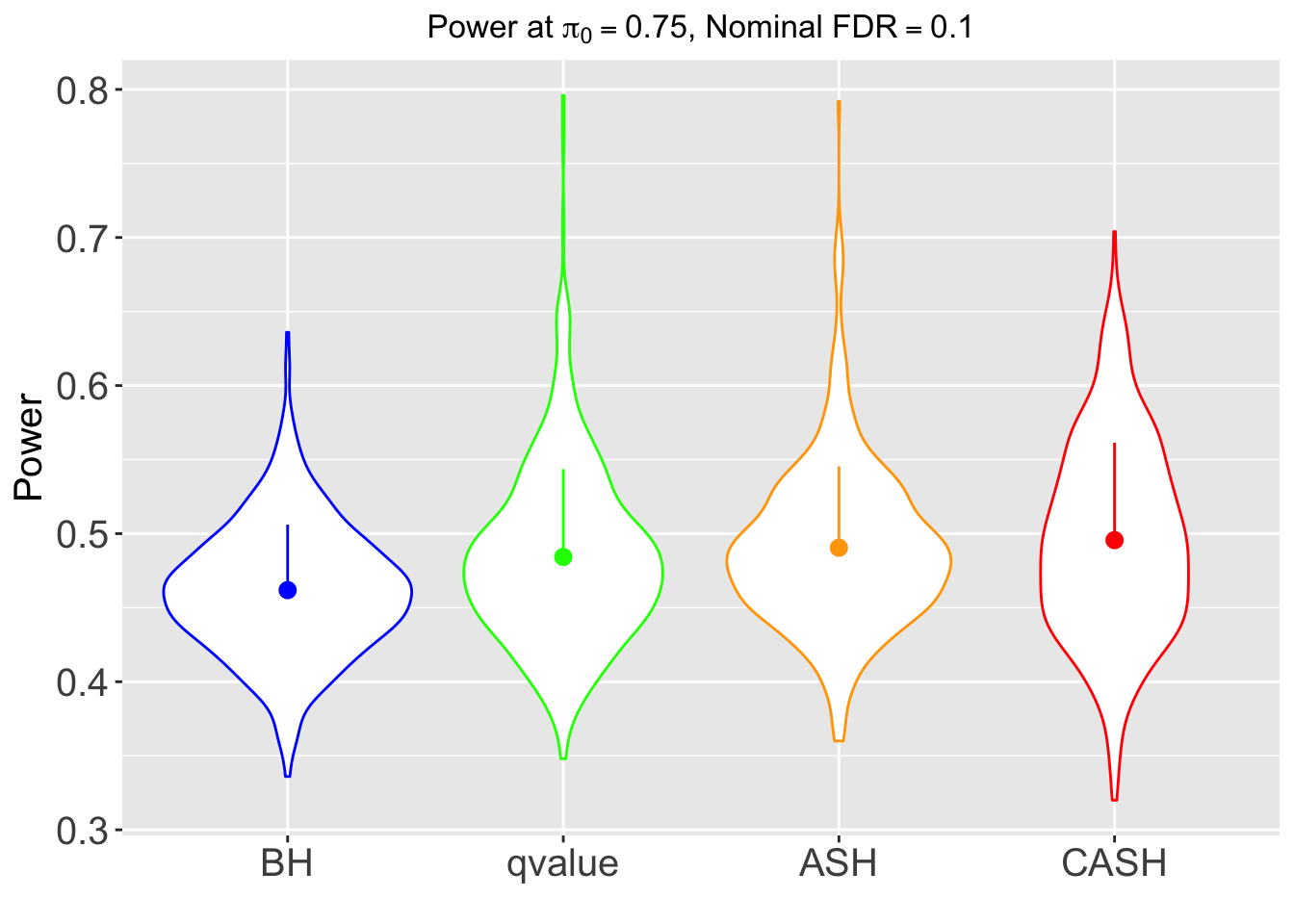
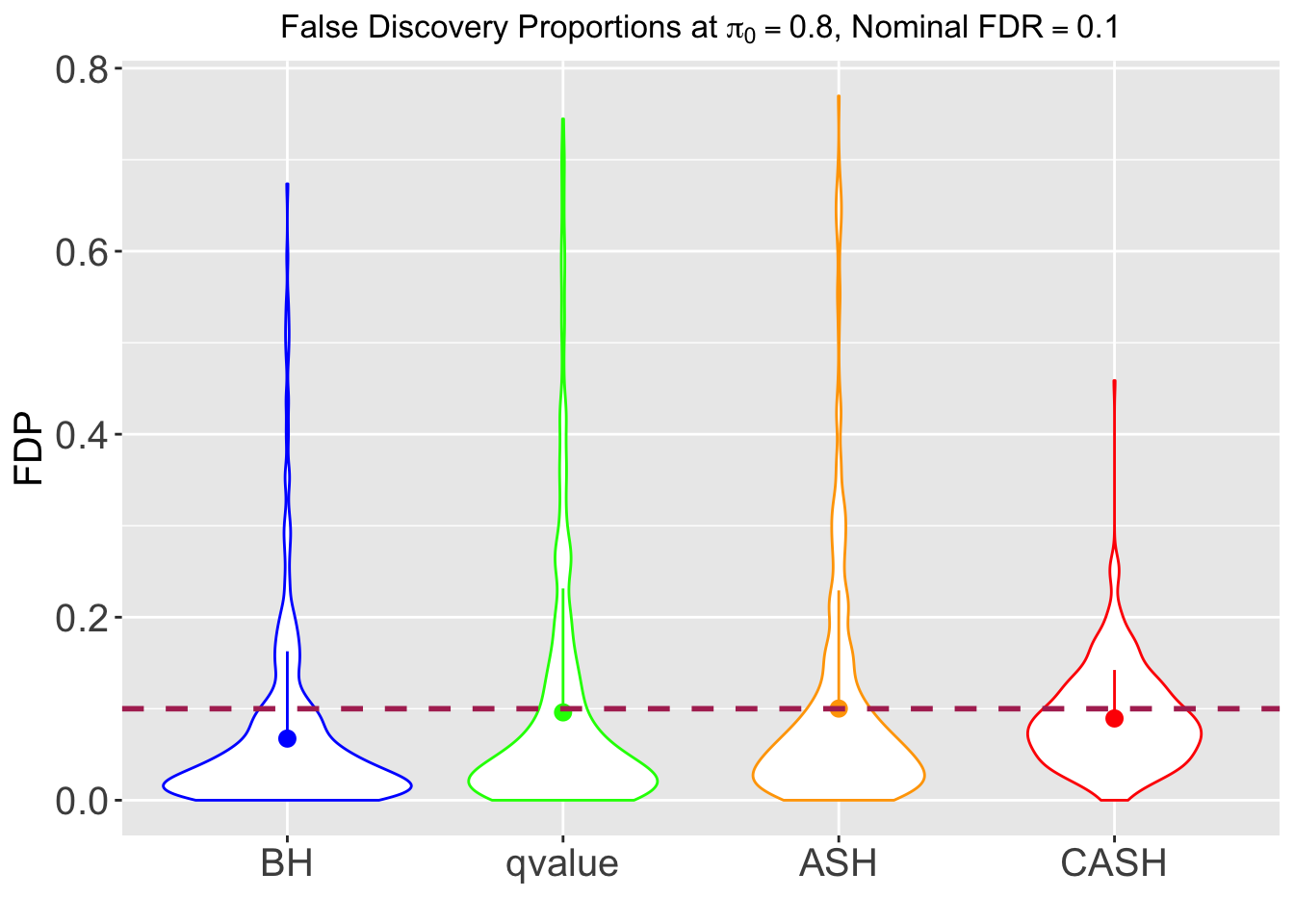
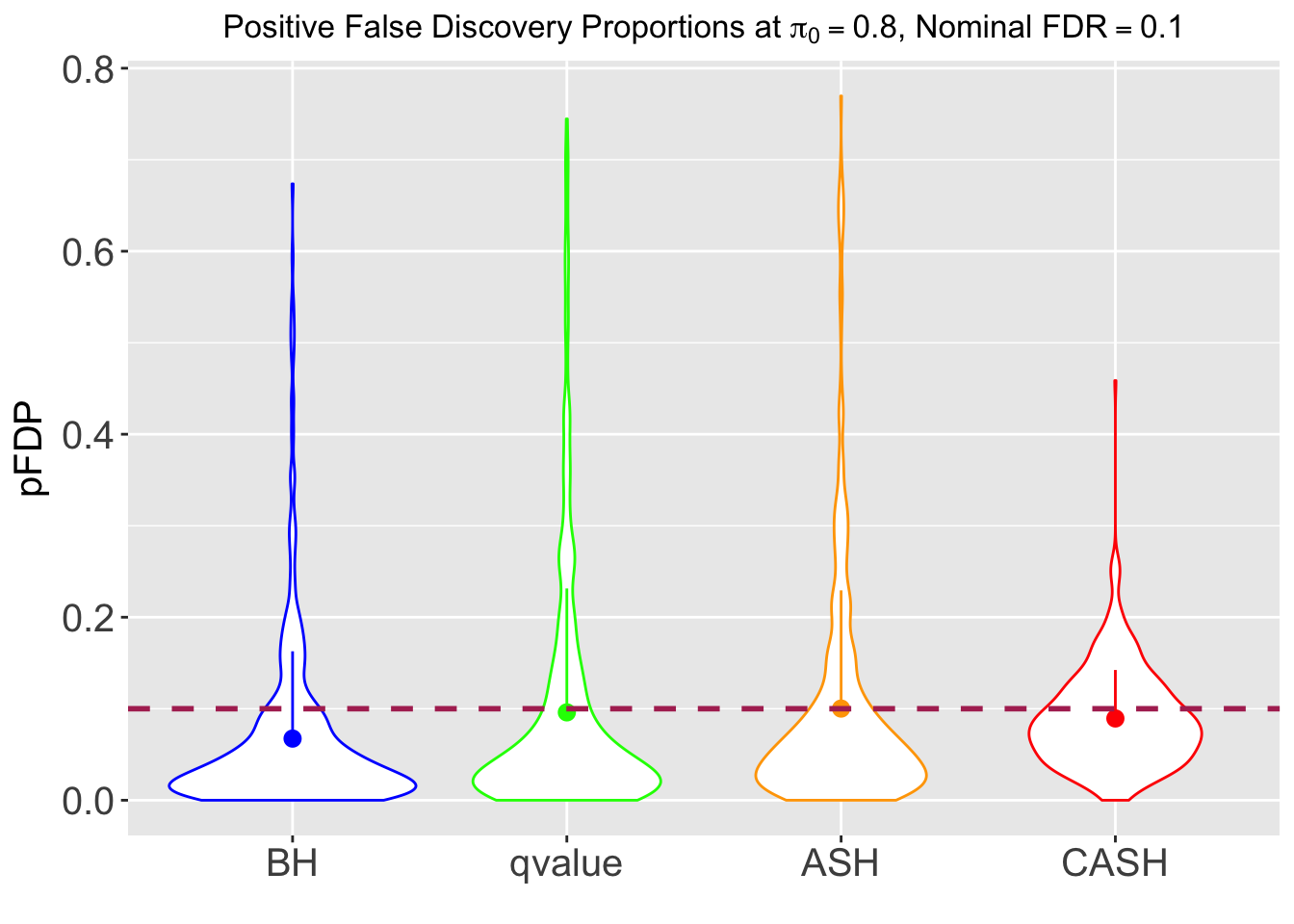
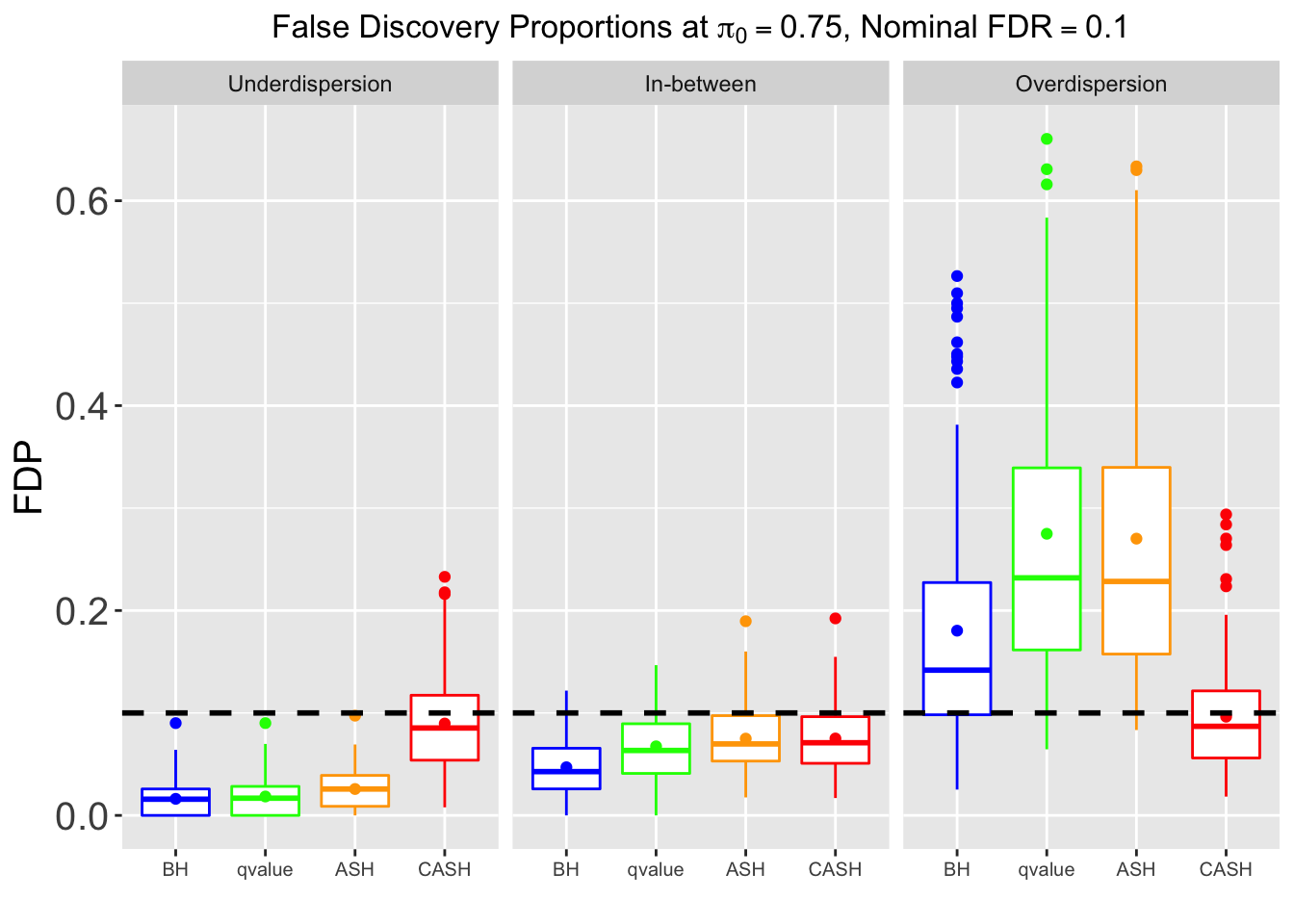
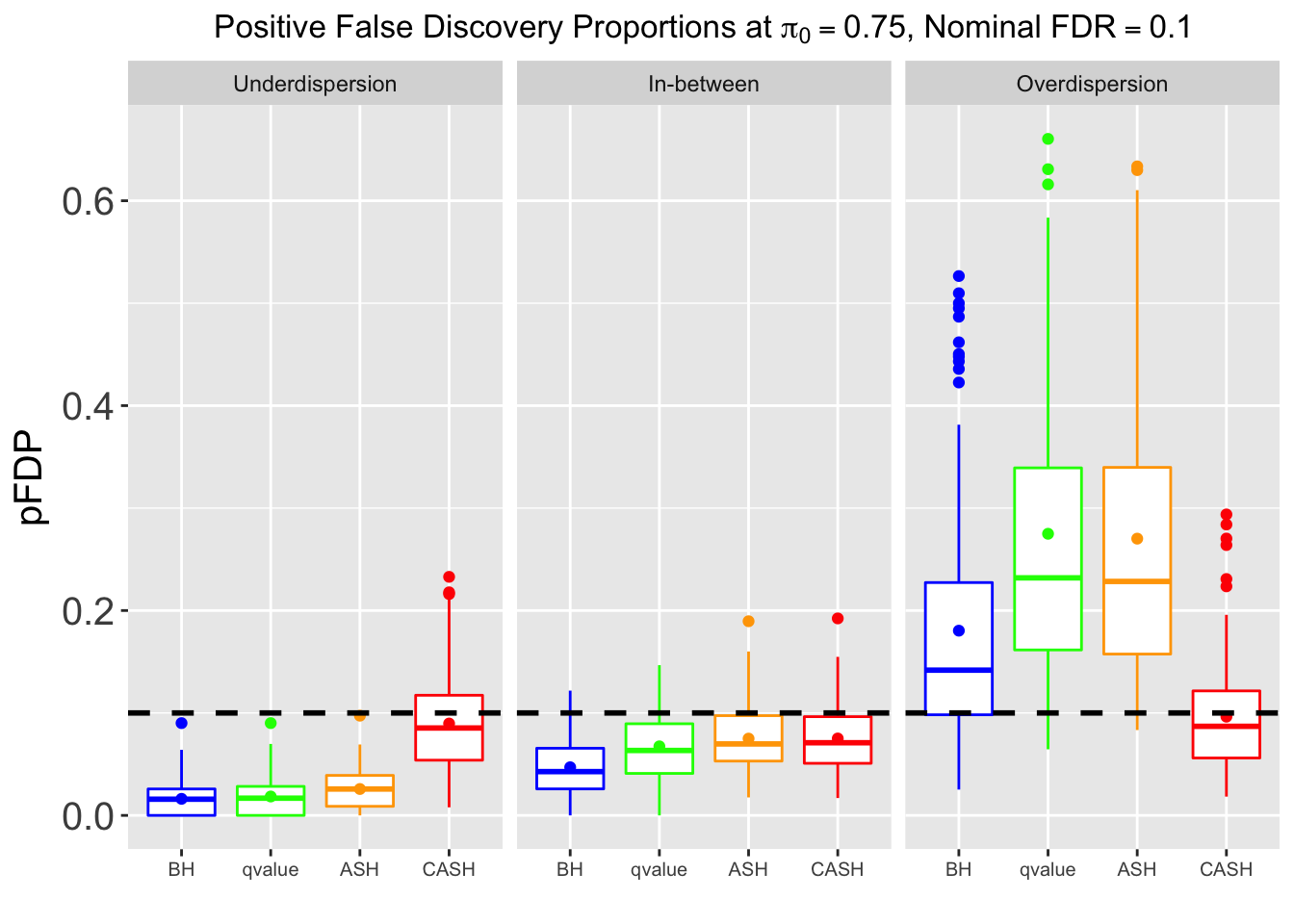
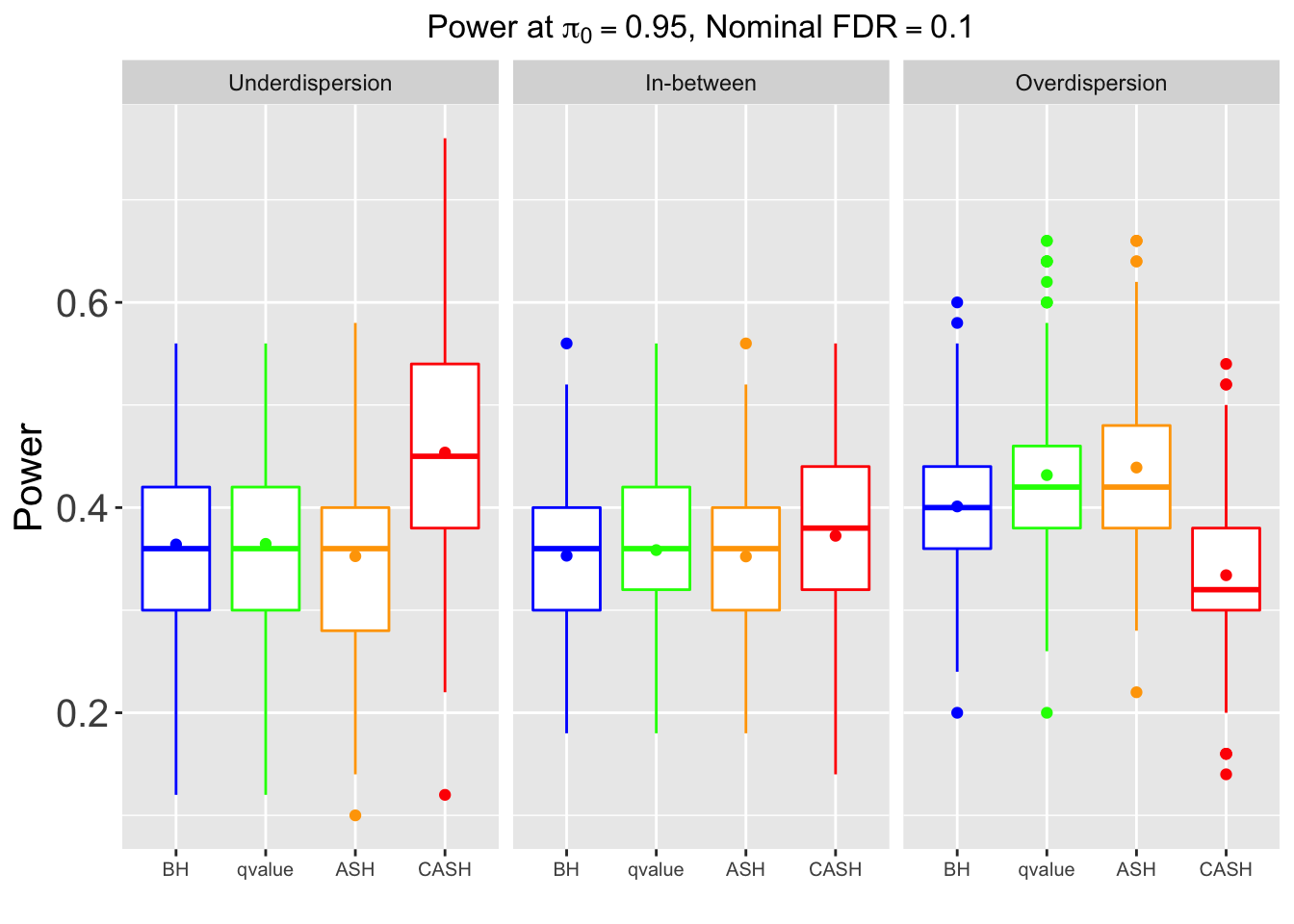
FDR, Power at each sparsity level at Nominal FDR = 0.1, separated by dispersion
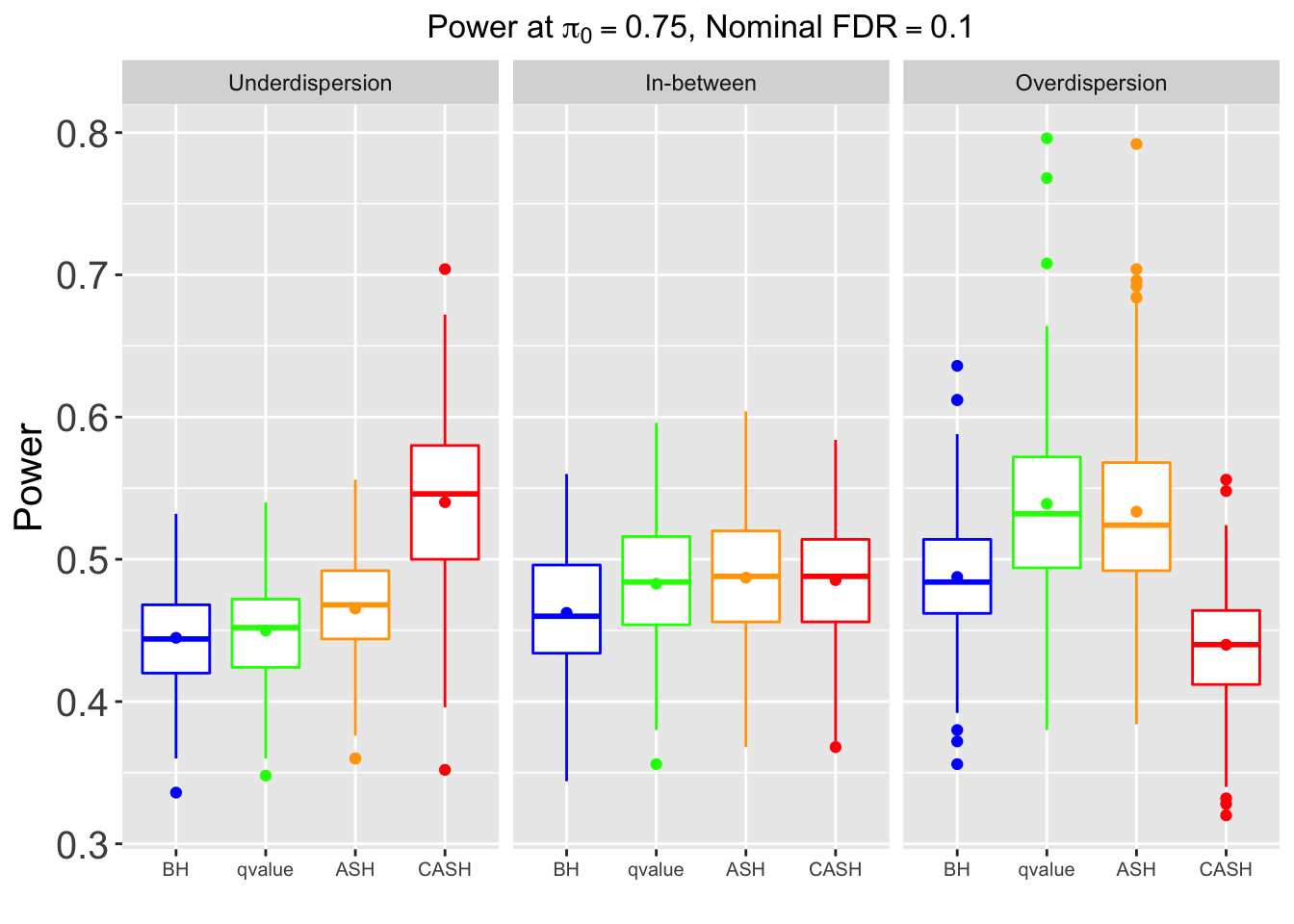
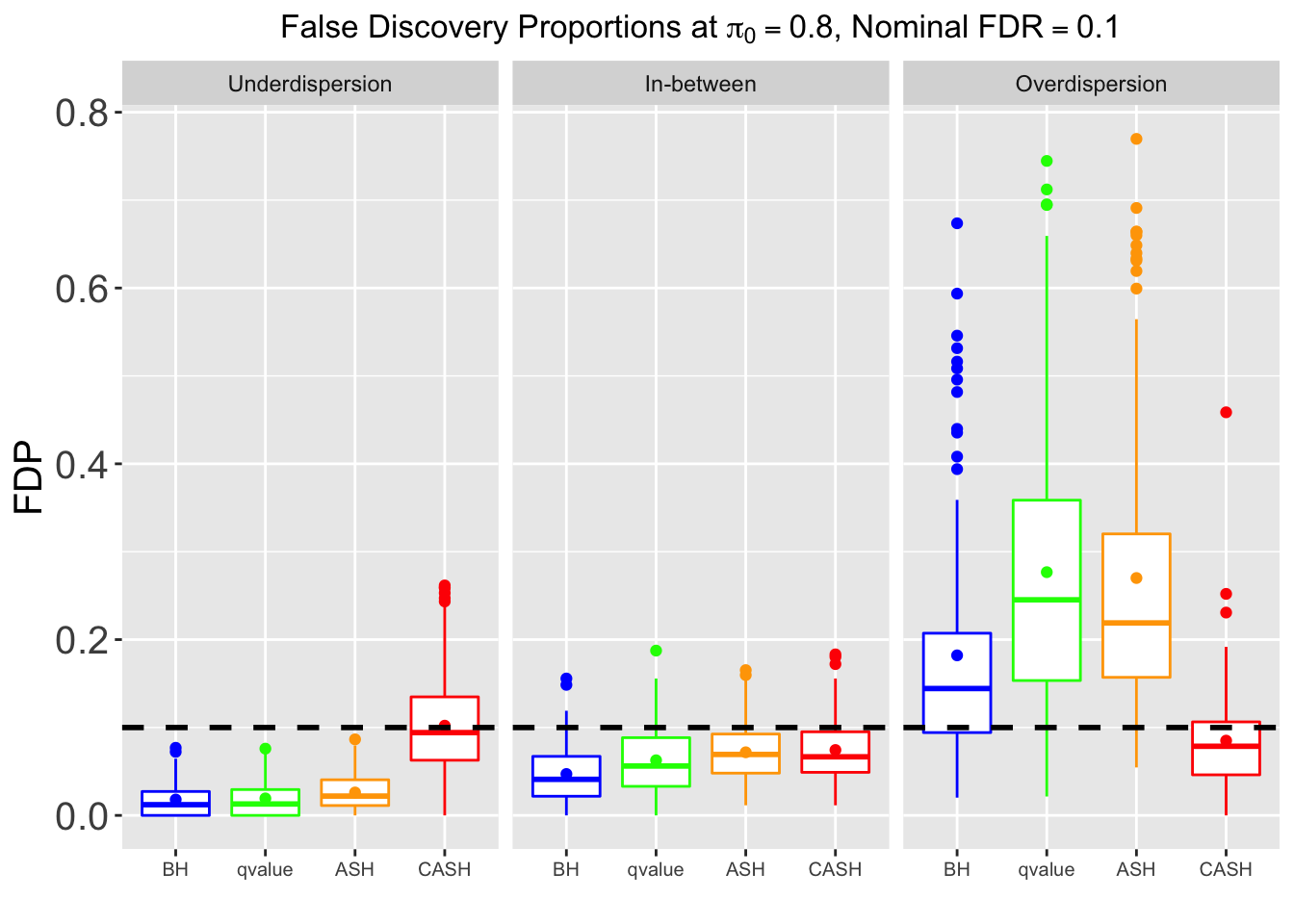
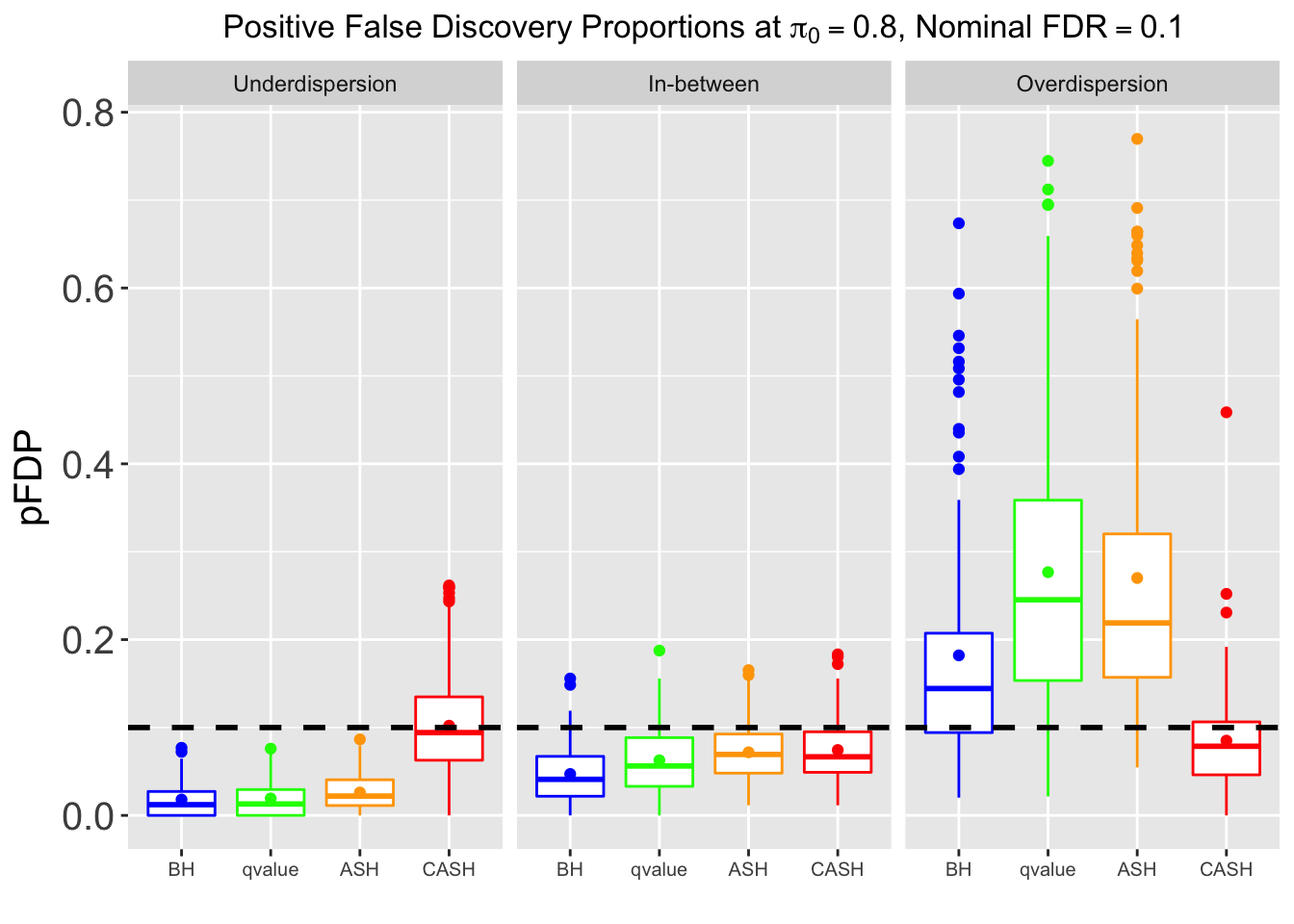
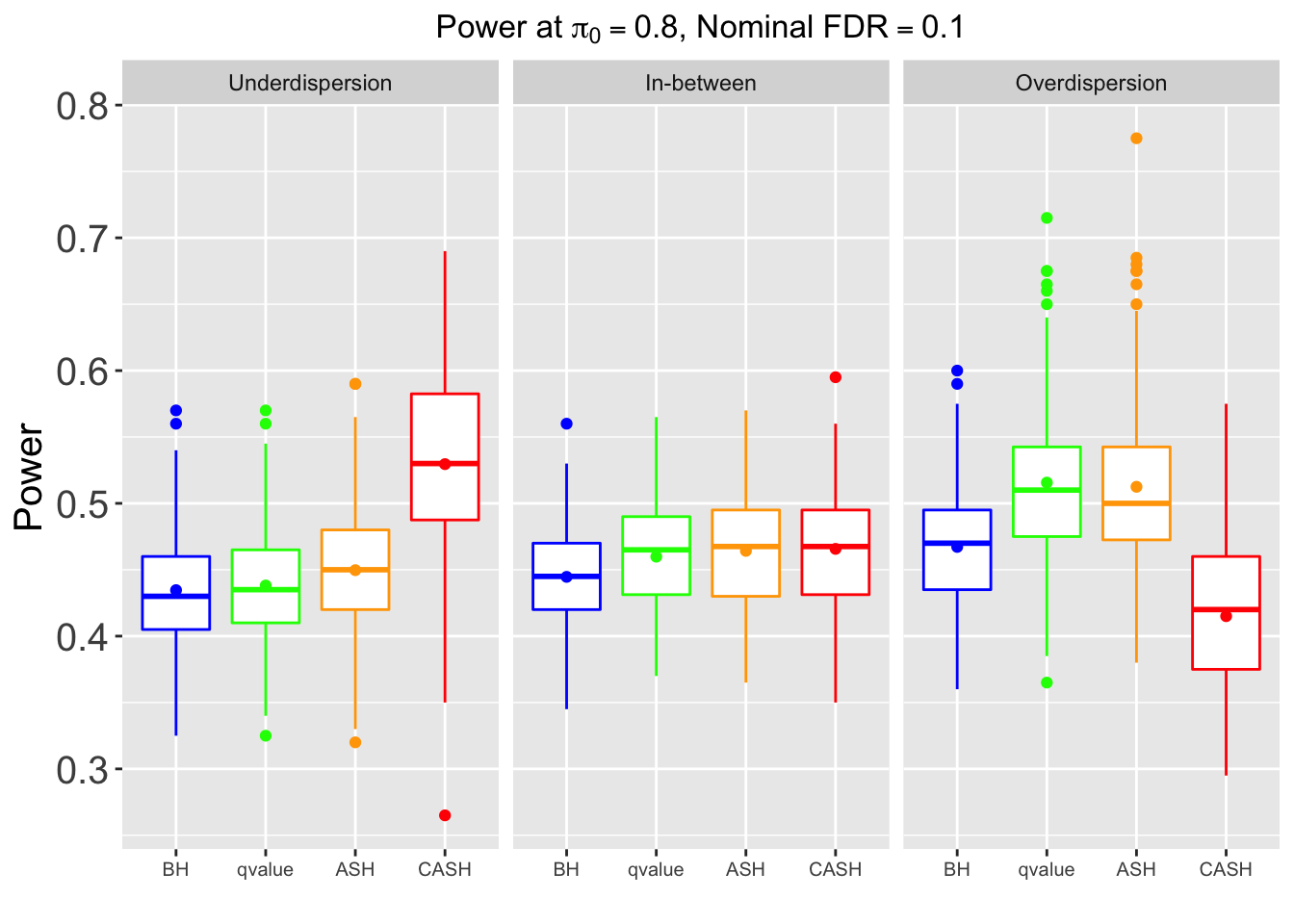
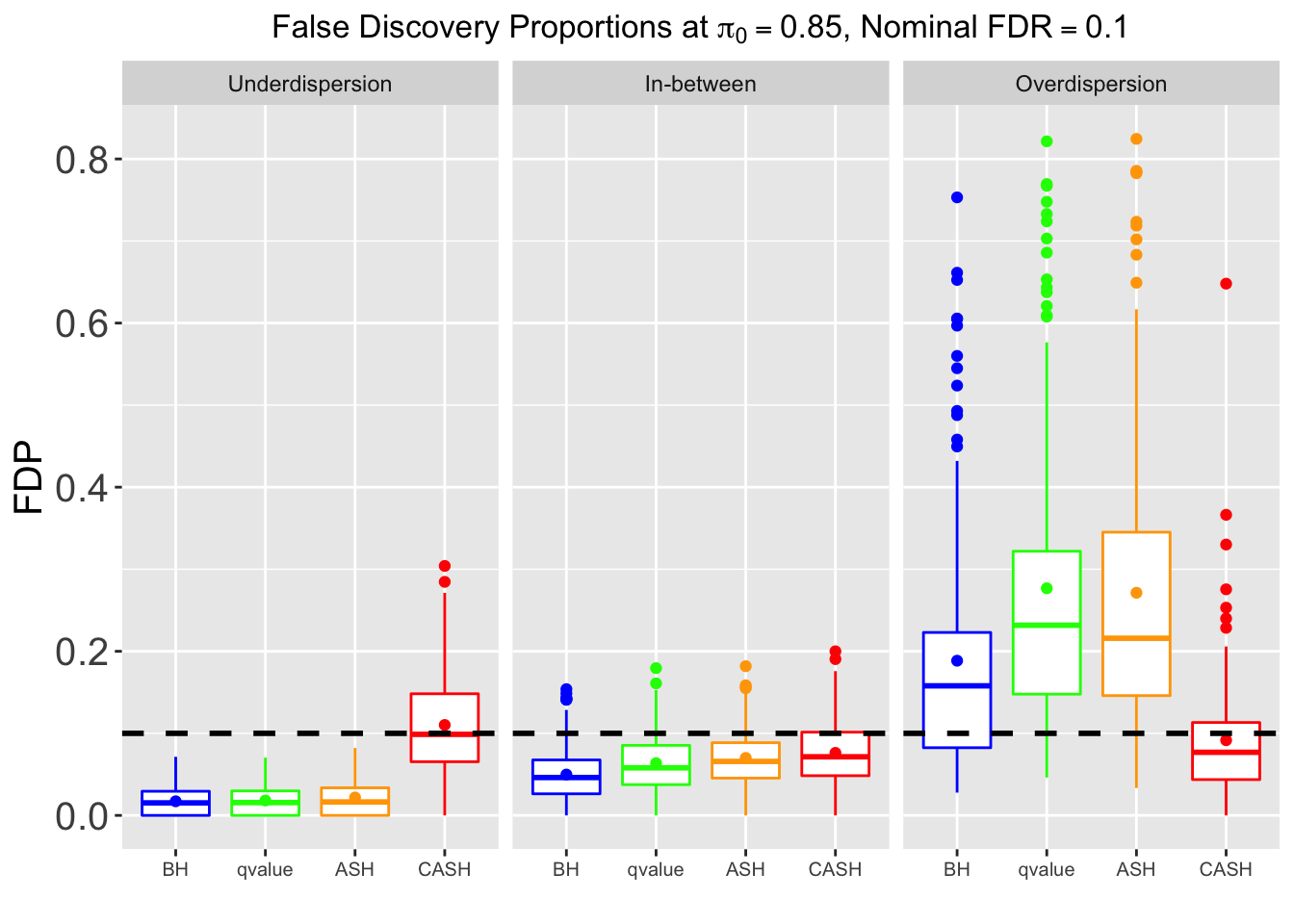
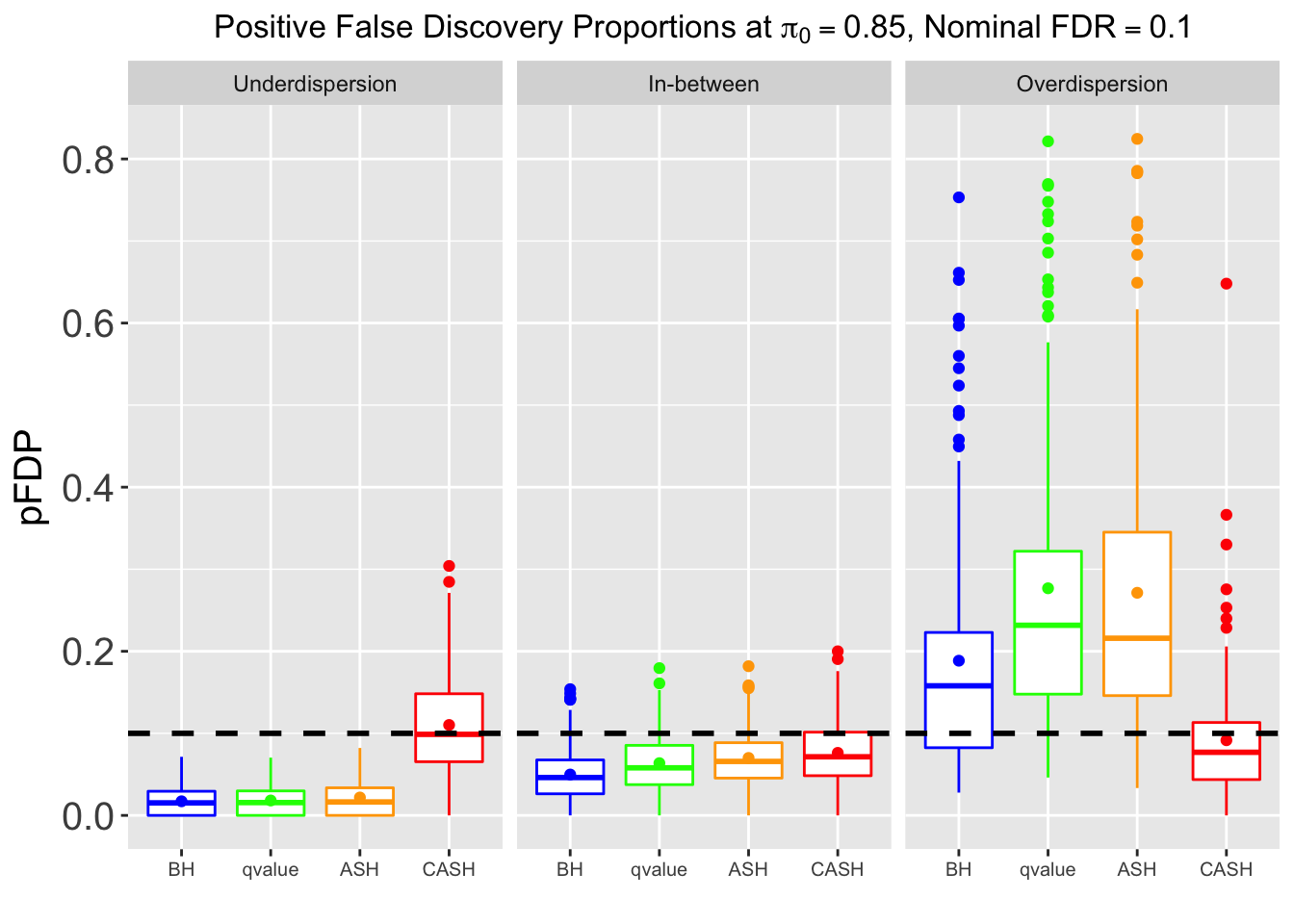
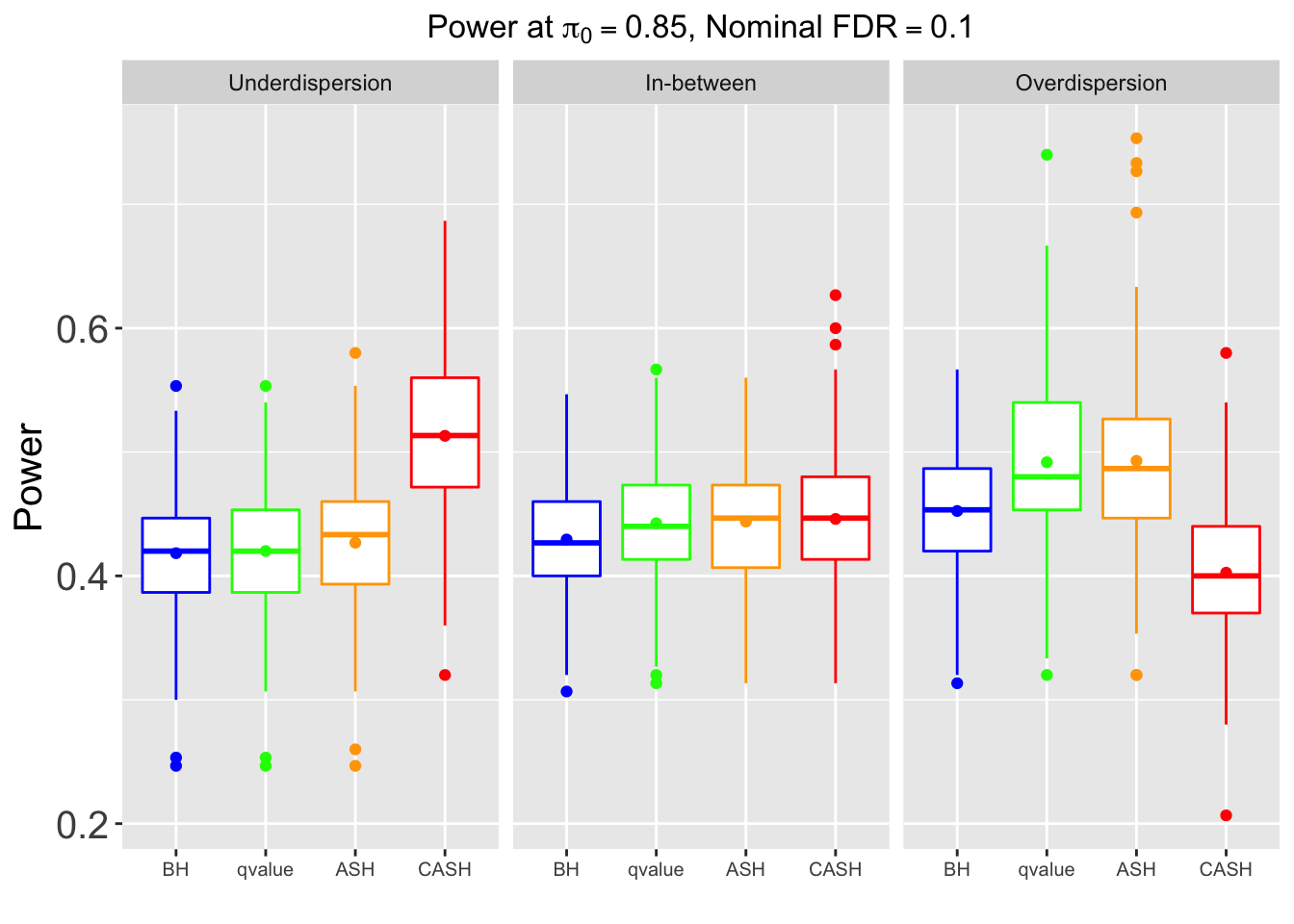
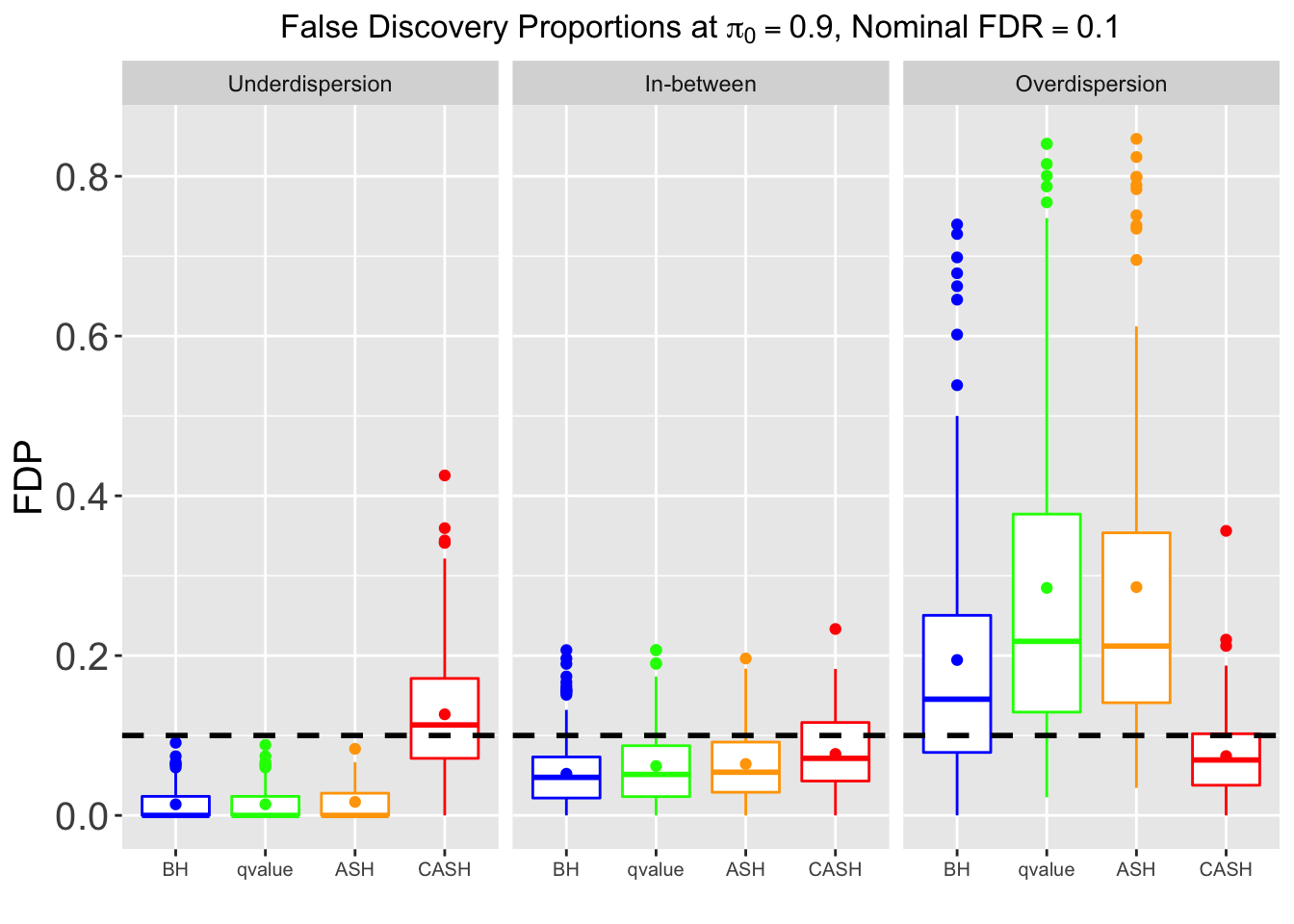
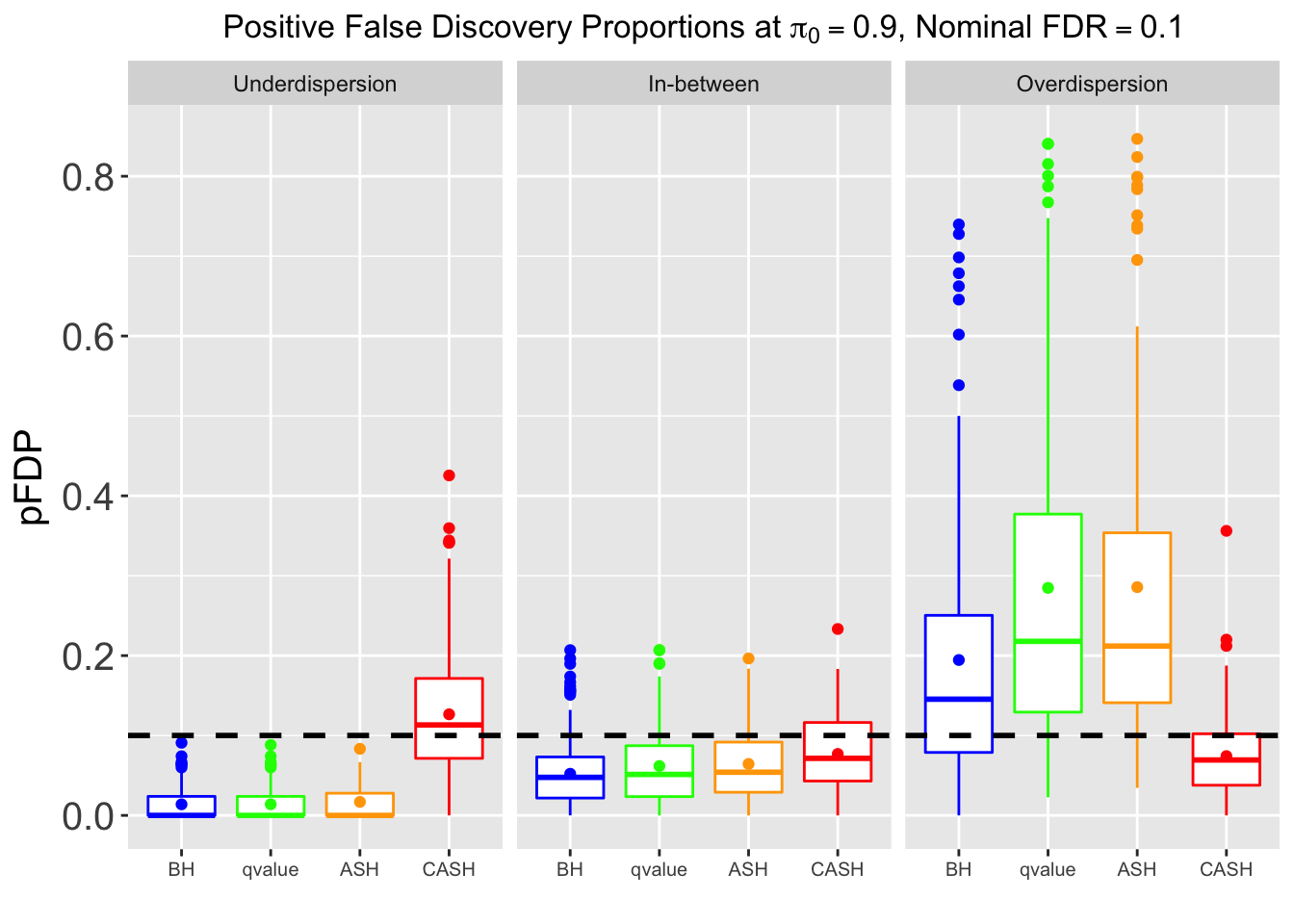
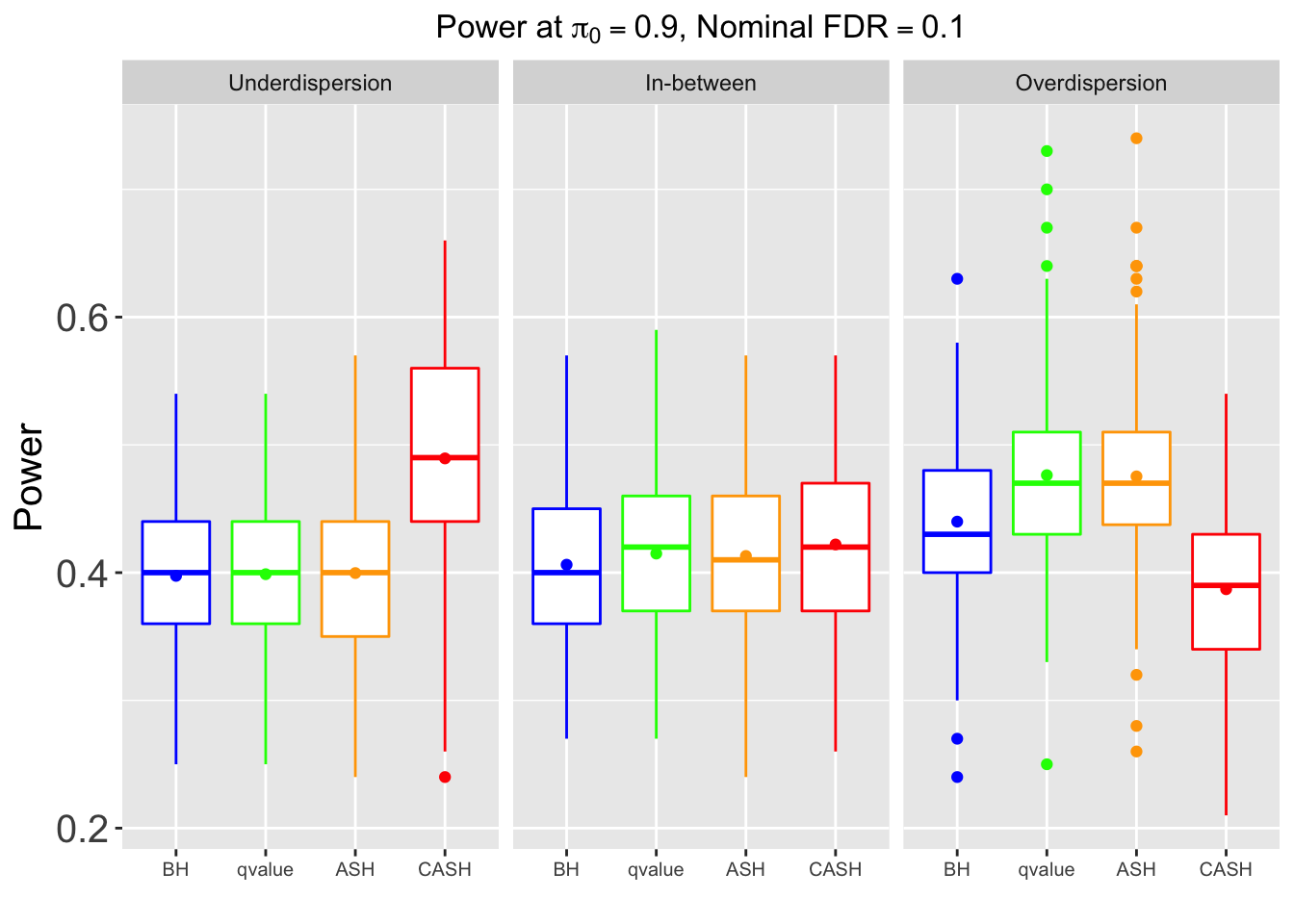
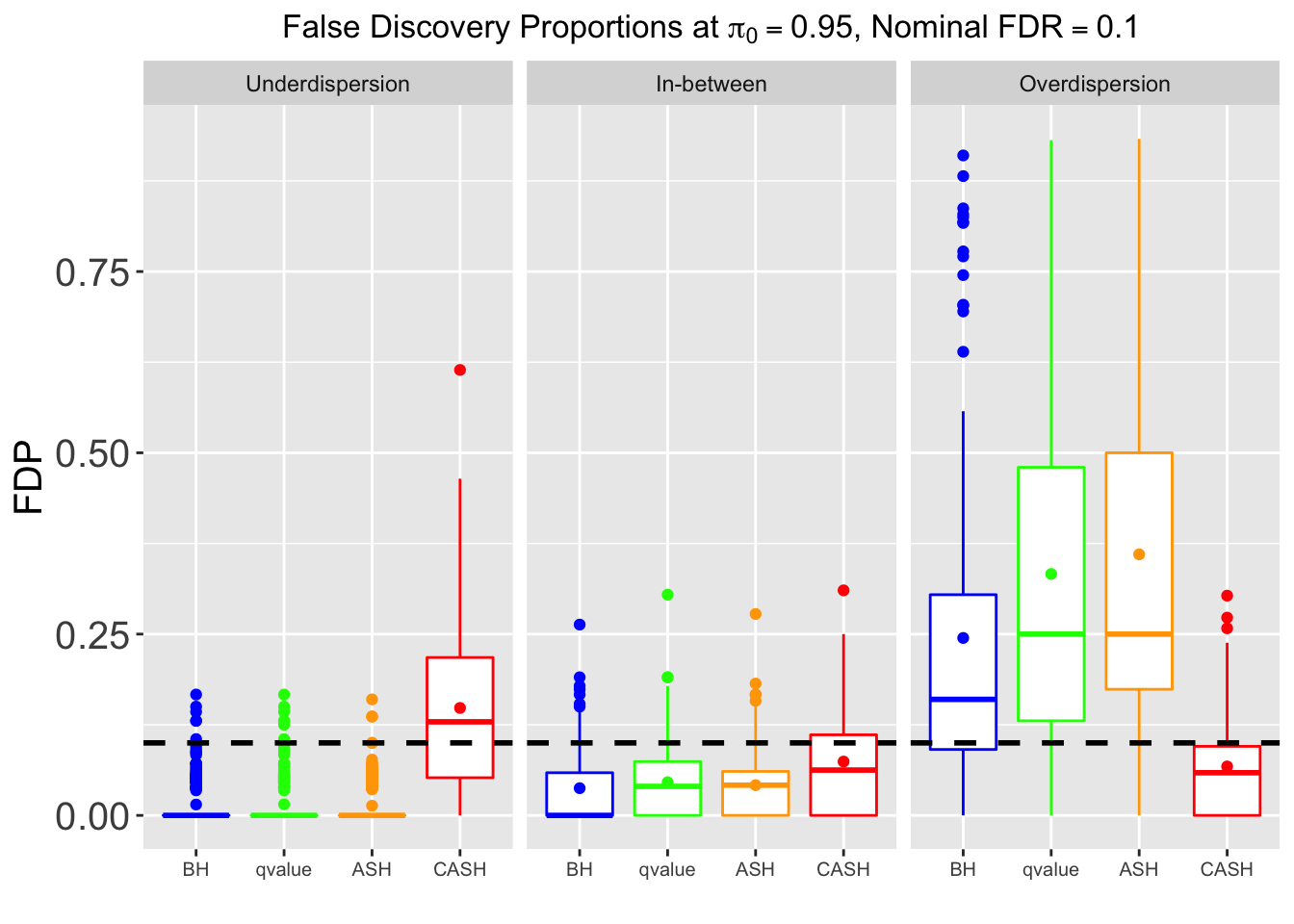
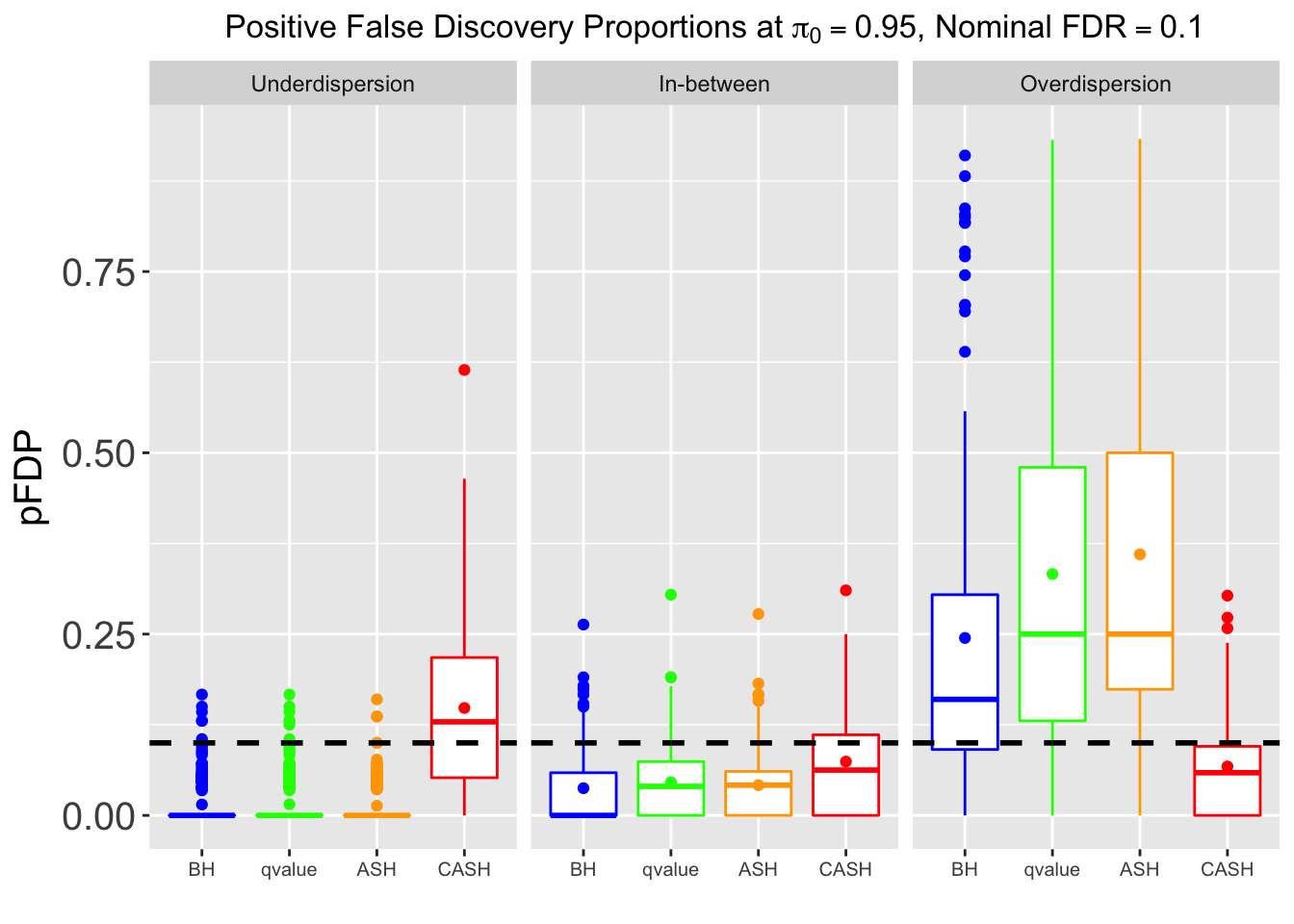
Many different \(\sigma_\beta\) and Gaussian deviatives
sigma.beta = 1 ; GD = 6 ; Constraints on w: TRUE

Simulated noise
sigma.beta = 1 ; GD = 6 ; Constraints on w: TRUE

sigma.beta = 2 ; GD = 6 ; Constraints on w: TRUE

sigma.beta = 3 ; GD = 6 ; Constraints on w: TRUE

sigma.beta = 4 ; GD = 6 ; Constraints on w: TRUE

sigma.beta = 5 ; GD = 6 ; Constraints on w: TRUE

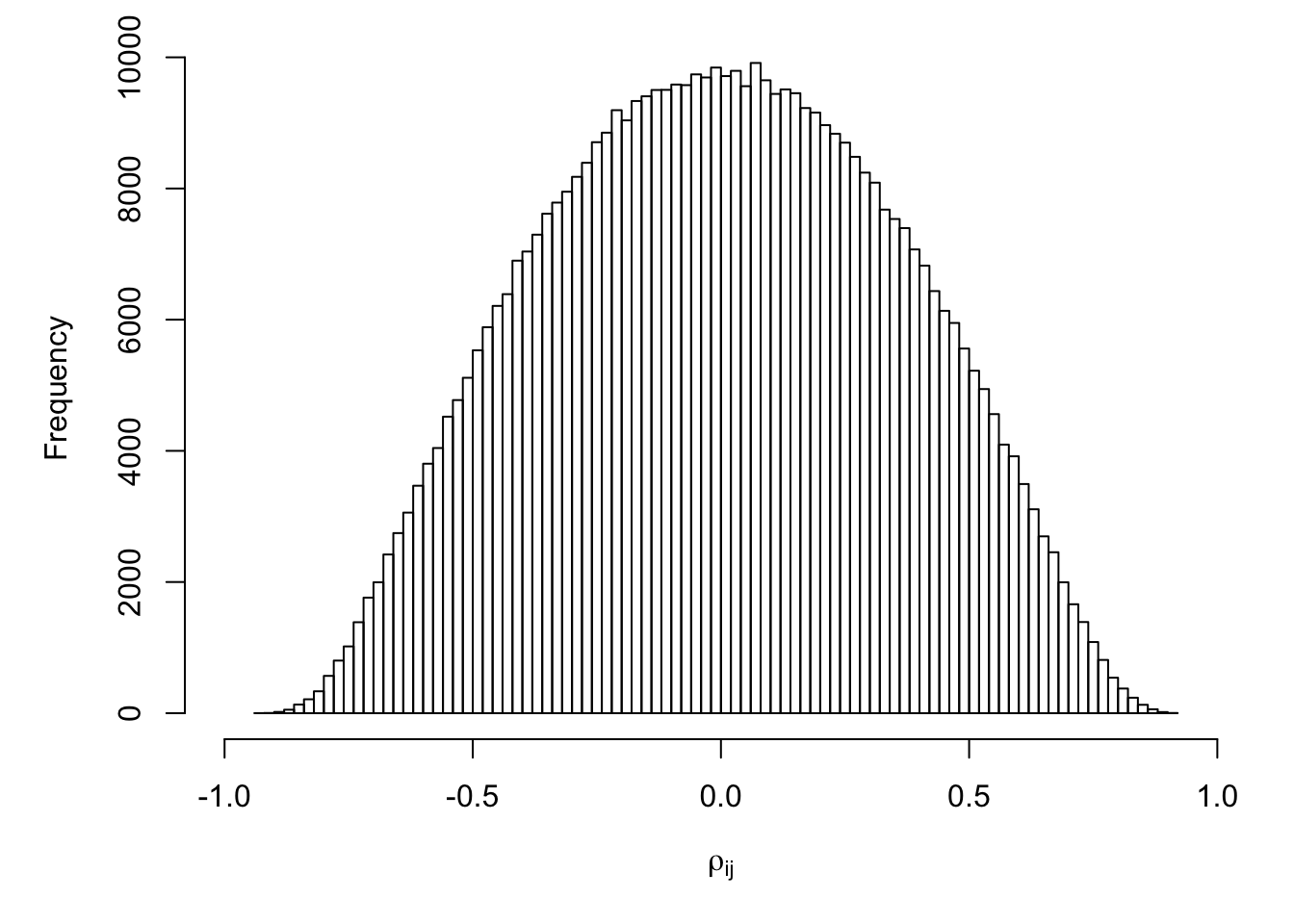
Distribution of pairwise correlations
set.seed(1)
z <- MASS::mvrnorm(n = 1, mu = rep(0, p), Sigma = Rho)histz <- hist(z, prob = TRUE, breaks = 20, xlab = "Z", main = "", xlim = c(-4, 4), ylim = c(0, 0.5))
lines(seq(-4, 4, by = 0.1), dnorm(seq(-4, 4, by = 0.1)), col = "blue")
legend("topright", lty = 1, col = "blue", "N(0, 1)")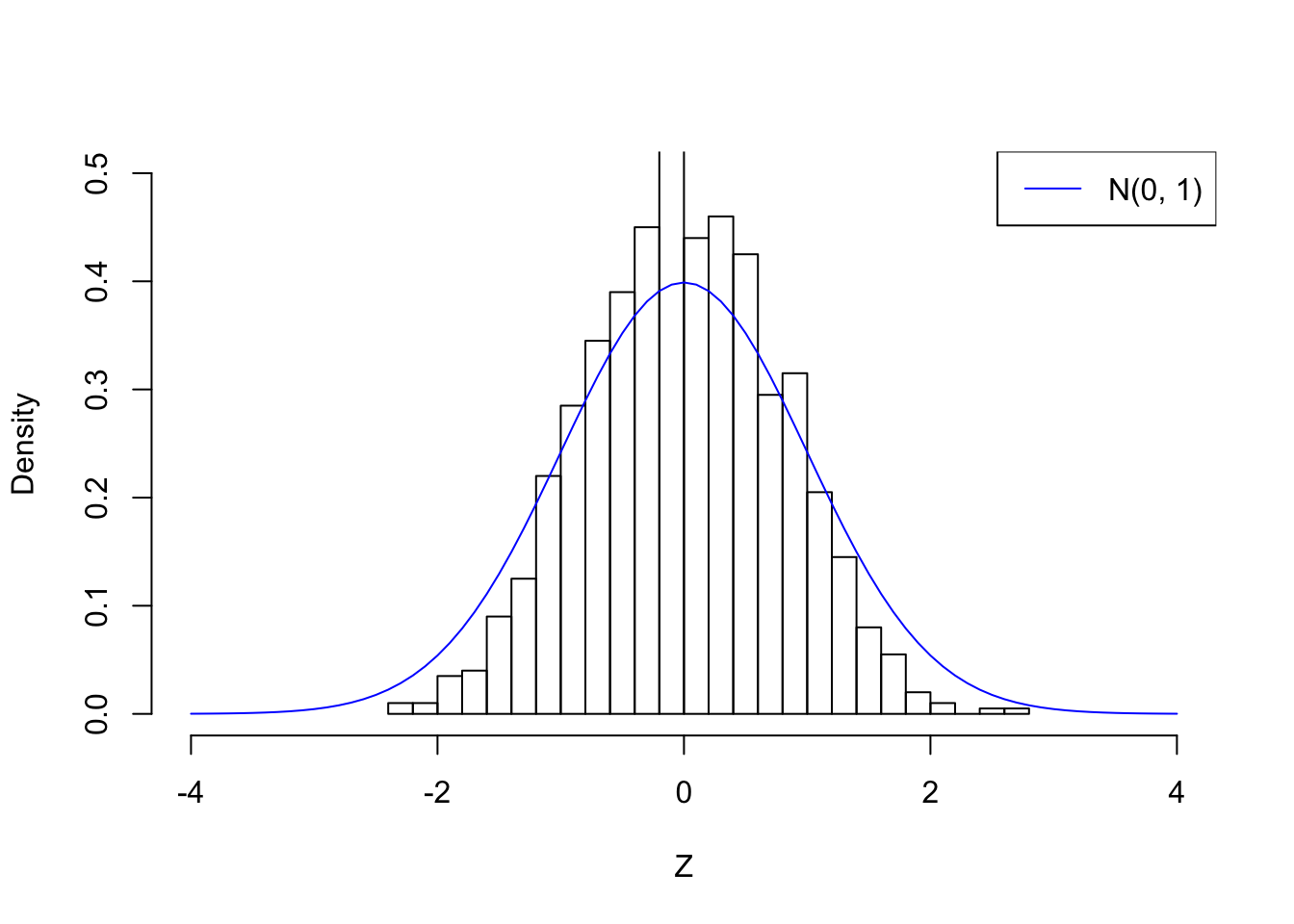
set.seed(7)
z <- MASS::mvrnorm(n = 1, mu = rep(0, p), Sigma = Rho)histz <- hist(z, prob = TRUE, breaks = 20, xlab = "Z", main = "", xlim = c(-4, 4), ylim = c(0, 0.5))
lines(seq(-4, 4, by = 0.1), dnorm(seq(-4, 4, by = 0.1)), col = "blue")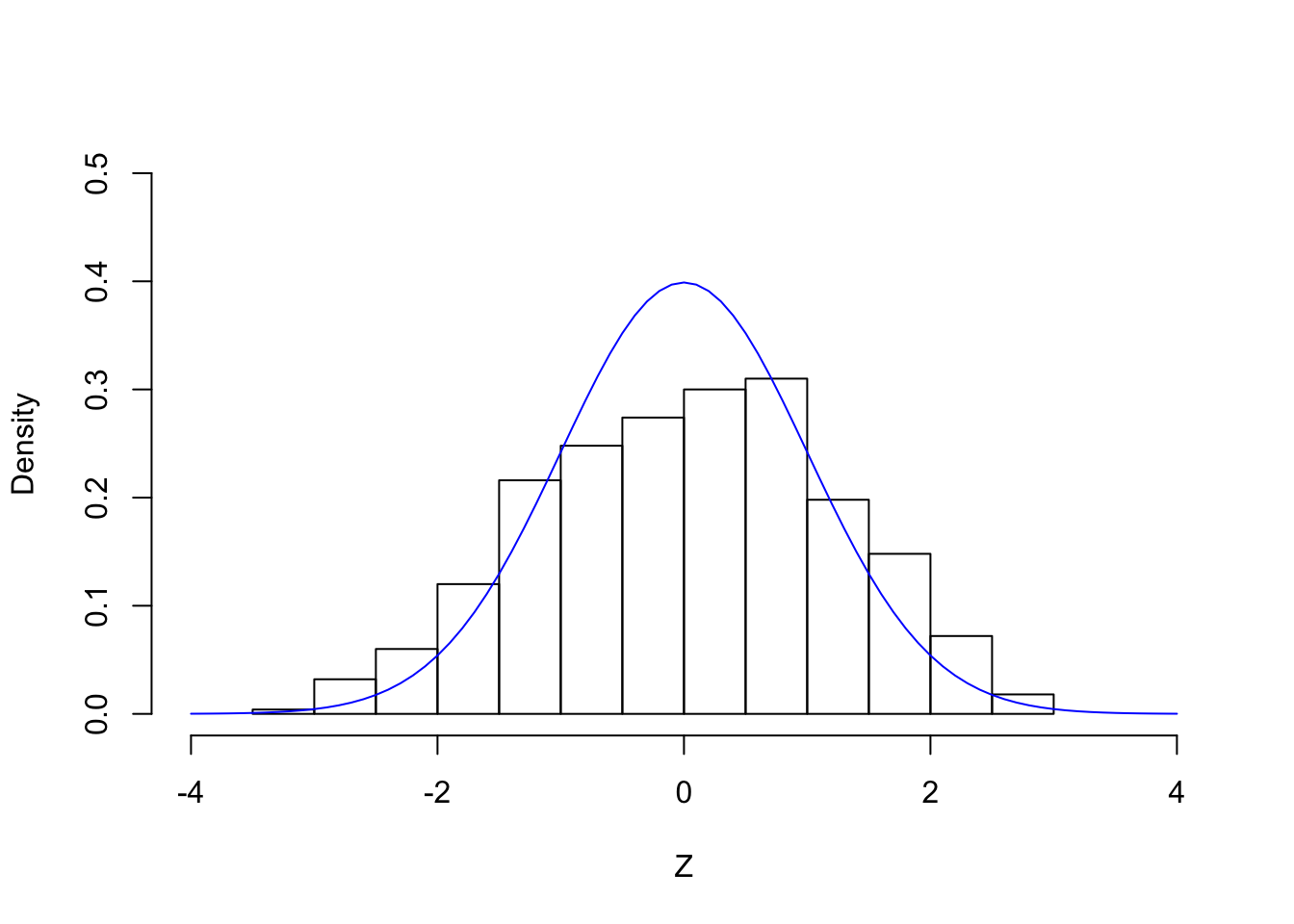
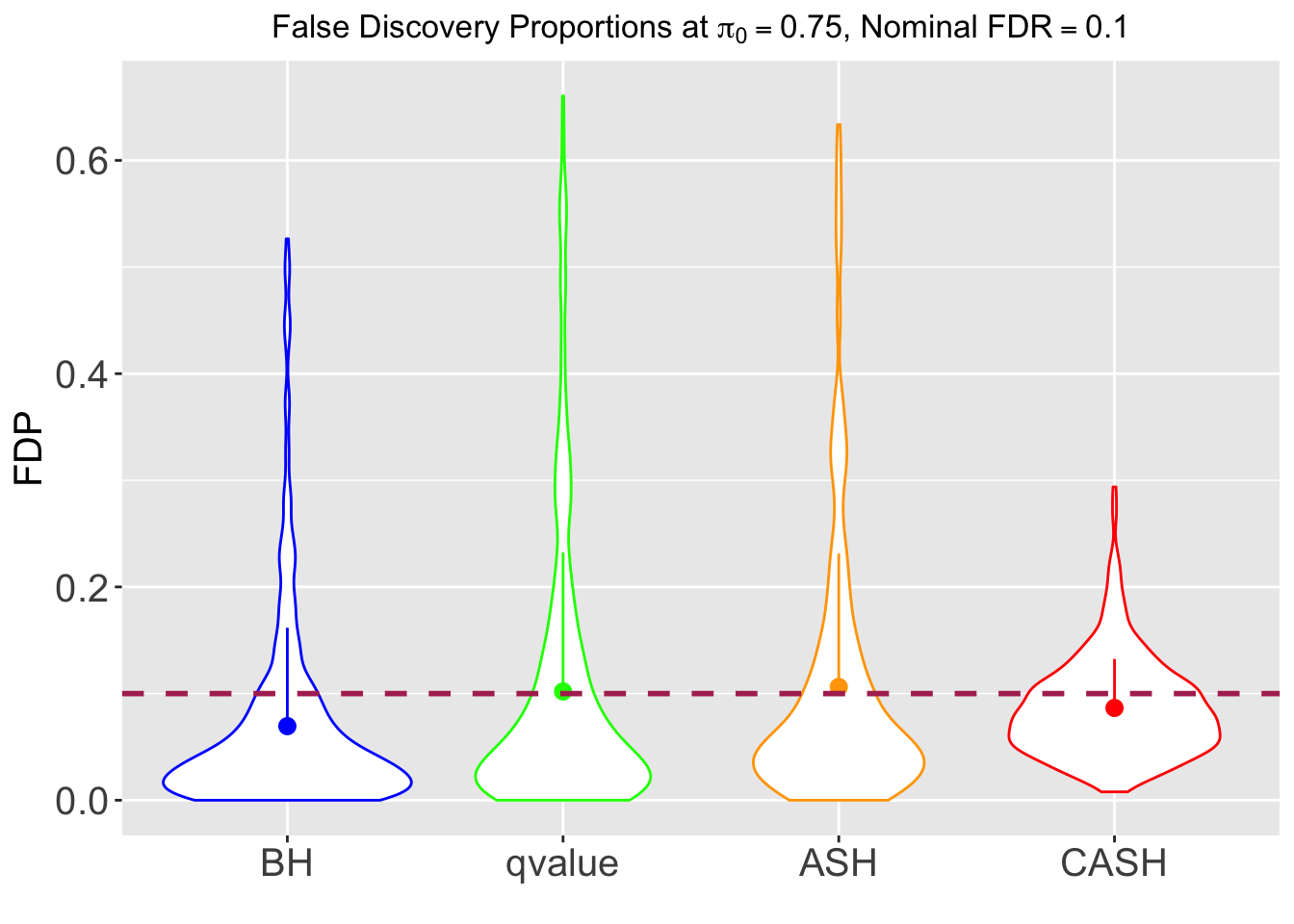
Variability of FDR and Power
FDP.array <- pFDP.array <- power.array <- array(0, dim = c(nsim, length(q.vec), length(method.name), length(pi0.vec)))
FDP.summary <- pFDP.summary <- power.summary <- array(0, dim = c(5, length(q.vec), length(method.name), length(pi0.vec)))
for (j in seq(length(pi0.vec))) {
for (k in seq(length(method.name))) {
for (i in seq(nsim)) {
FDP.array[i, , k, j] <- sapply(q.vec, FDP, qvalue = qvalue.pi0.list[[j]][[i]][, k + 1], beta = qvalue.pi0.list[[j]][[i]][, 1])
pFDP.array[i, , k, j] <- sapply(q.vec, pFDP, qvalue = qvalue.pi0.list[[j]][[i]][, k + 1], beta = qvalue.pi0.list[[j]][[i]][, 1])
power.array[i, , k, j] <- sapply(q.vec, power, qvalue = qvalue.pi0.list[[j]][[i]][, k + 1], beta = qvalue.pi0.list[[j]][[i]][, 1])
}
FDP.summary[, , k, j] <- rbind(
avg <- colMeans(FDP.array[, , k, j]),
sd <- apply(FDP.array[, , k, j], 2, sd),
n <- colSums(!is.na(FDP.array[, , k, j])),
q975 <- apply(FDP.array[, , k, j], 2, quantile, probs = 0.975),
q025 <- apply(FDP.array[, , k, j], 2, quantile, probs = 0.025)
)
pFDP.summary[, , k, j] <- rbind(
avg <- colMeans(pFDP.array[, , k, j], na.rm = TRUE),
sd <- apply(pFDP.array[, , k, j], 2, sd, na.rm = TRUE),
n <- colSums(!is.na(pFDP.array[, , k, j])),
q975 <- apply(pFDP.array[, , k, j], 2, quantile, probs = 0.975, na.rm = TRUE),
q025 <- apply(pFDP.array[, , k, j], 2, quantile, probs = 0.025, na.rm = TRUE)
)
power.summary[, , k, j] <- rbind(
avg <- colMeans(power.array[, , k, j]),
sd <- apply(power.array[, , k, j], 2, sd),
n <- colSums(!is.na(power.array[, , k, j])),
q975 <- apply(power.array[, , k, j], 2, quantile, probs = 0.975),
q025 <- apply(power.array[, , k, j], 2, quantile, probs = 0.025)
)
}
}
for (j in seq(length(pi0.vec))) {
for (q in c(0.1, 0.05)) {
sd.z <- sapply(z.pi0.list[[j]], sd)
Noise <- cut(sd.z, breaks = c(0, z.under, z.over, Inf), labels = c("Deflated Error", "In-between", "Inflated Error"))
FDR.plot <- ggplot(data = melt(cbind(Noise, as.data.frame(FDP.array[, which(round(q.vec, 4) == q), , j])), id.vars = "Noise"),
aes(x = variable, y = value, col = variable)) +
geom_boxplot() +
stat_summary(fun.y = mean, geom = "point", shape = 13, size = 3) +
scale_color_manual(values = method.col) +
facet_wrap(~Noise) +
geom_hline(yintercept = q, col = "black", linetype = "dashed", size = 1) +
scale_x_discrete(labels = method.name) +
labs(x = "", y = "FDP") +
theme(legend.position = "none", plot.title = element_text(hjust = 0.5, size = 12.5), axis.title.y = element_text(size = 15), axis.text.x = element_text(size = 7.5), axis.text.y = element_text(size = 15))
# pFDR.plot <- ggplot(data = melt(cbind(Noise, as.data.frame(pFDP.array[, which(q.vec == q), , j])), id.vars = "Noise"),
# aes(x = variable, y = value, col = variable)) +
# geom_boxplot(na.rm = TRUE) +
# stat_summary(fun.y = mean, geom = "point") +
# scale_color_manual(values = method.col) +
# facet_wrap(~Noise) +
# geom_hline(yintercept = q, col = "black", linetype = "dashed", size = 1) +
# scale_x_discrete(labels = method.name) +
# labs(x = "", y = "Positive FDP") +
# theme(legend.position = "none", plot.title = element_text(hjust = 0.5, size = 12.5), axis.title.y = element_text(size = 15), axis.text.x = element_text(size = 7.5), axis.text.y = element_text(size = 15))
power.plot <- ggplot(data = melt(cbind(Noise, as.data.frame(power.array[, which(round(q.vec, 4) == q), , j])), id.vars = "Noise"),
aes(x = variable, y = value, col = variable)) +
geom_boxplot() +
stat_summary(fun.y = mean, geom = "point", shape = 8, size = 2) +
scale_color_manual(values = method.col) +
facet_wrap(~Noise) +
scale_x_discrete(labels = method.name) +
labs(x = "", y = "TPP") +
theme(legend.position = "none", plot.title = element_text(hjust = 0.5, size = 12.5), axis.title.y = element_text(size = 15), axis.text.x = element_text(size = 7.5), axis.text.y = element_text(size = 15))
grid.arrange(FDR.plot, power.plot
#, top = textGrob(bquote(paste(pi[0] == .(pi0.vec[j]), "; ", sigma[beta] == .(sigma.beta), "; ", .(L), "GDs; ", .(ifelse(w.lambda == 0, "No Constraints", "With Constraints")), "; ", q == .(q))))
)
# pdf(paste("../output/fig/sim_cash_2_", sigma.beta, L, w.lambda, pi0.vec[j], "plot.pdf", sep = "_"), onefile = TRUE)
# grid.arrange(FDR.plot, power.plot, top = textGrob(bquote(paste(pi[0] == .(pi0.vec[j]), "; ", sigma[beta] == .(sigma.beta), "; ", .(L), "GDs; ", .(ifelse(w.lambda == 0, "No Constraints", "With Constraints")), "; ", q == .(q)))))
# dev.off()
}
}
Large-scale simulation with real data
r <- readRDS("../data/liver.rds")top_genes_index = function (g, X) {
return(order(rowSums(X), decreasing = TRUE)[1 : g])
}
lcpm = function (r) {
R = colSums(r)
t(log2(((t(r) + 0.5) / (R + 1)) * 10^6))
}nsamp <- 5
ngene <- 1e4
nsim <- 1000
pi0.vec <- 0.9Y = lcpm(r)
subset = top_genes_index(ngene, Y)
r = r[subset,]z.over <- 1.025
z.under <- 0.975
method.name <- c("BH", "qvalue", "locfdr", "ASH", "CASH")FDP.array <- pFDP.array <- power.array <- array(0, dim = c(nsim, length(q.vec), length(method.name), length(pi0.vec)))
FDP.summary <- pFDP.summary <- power.summary <- array(0, dim = c(5, length(q.vec), length(method.name), length(pi0.vec)))
for (j in seq(length(pi0.vec))) {
for (k in seq(length(method.name))) {
for (i in seq(nsim)) {
FDP.array[i, , k, j] <- sapply(q.vec, FDP, qvalue = qvalue.pi0.list[[j]][[i]][, k + 1], beta = qvalue.pi0.list[[j]][[i]][, 1])
pFDP.array[i, , k, j] <- sapply(q.vec, pFDP, qvalue = qvalue.pi0.list[[j]][[i]][, k + 1], beta = qvalue.pi0.list[[j]][[i]][, 1])
power.array[i, , k, j] <- sapply(q.vec, power, qvalue = qvalue.pi0.list[[j]][[i]][, k + 1], beta = qvalue.pi0.list[[j]][[i]][, 1])
}
FDP.summary[, , k, j] <- rbind(
avg <- colMeans(FDP.array[, , k, j]),
sd <- apply(FDP.array[, , k, j], 2, sd),
n <- colSums(!is.na(FDP.array[, , k, j])),
q975 <- apply(FDP.array[, , k, j], 2, quantile, probs = 0.975),
q025 <- apply(FDP.array[, , k, j], 2, quantile, probs = 0.025)
)
pFDP.summary[, , k, j] <- rbind(
avg <- colMeans(pFDP.array[, , k, j], na.rm = TRUE),
sd <- apply(pFDP.array[, , k, j], 2, sd, na.rm = TRUE),
n <- colSums(!is.na(pFDP.array[, , k, j])),
q975 <- apply(pFDP.array[, , k, j], 2, quantile, probs = 0.975, na.rm = TRUE),
q025 <- apply(pFDP.array[, , k, j], 2, quantile, probs = 0.025, na.rm = TRUE)
)
power.summary[, , k, j] <- rbind(
avg <- colMeans(power.array[, , k, j]),
sd <- apply(power.array[, , k, j], 2, sd),
n <- colSums(!is.na(power.array[, , k, j])),
q975 <- apply(power.array[, , k, j], 2, quantile, probs = 0.975),
q025 <- apply(power.array[, , k, j], 2, quantile, probs = 0.025)
)
}
}
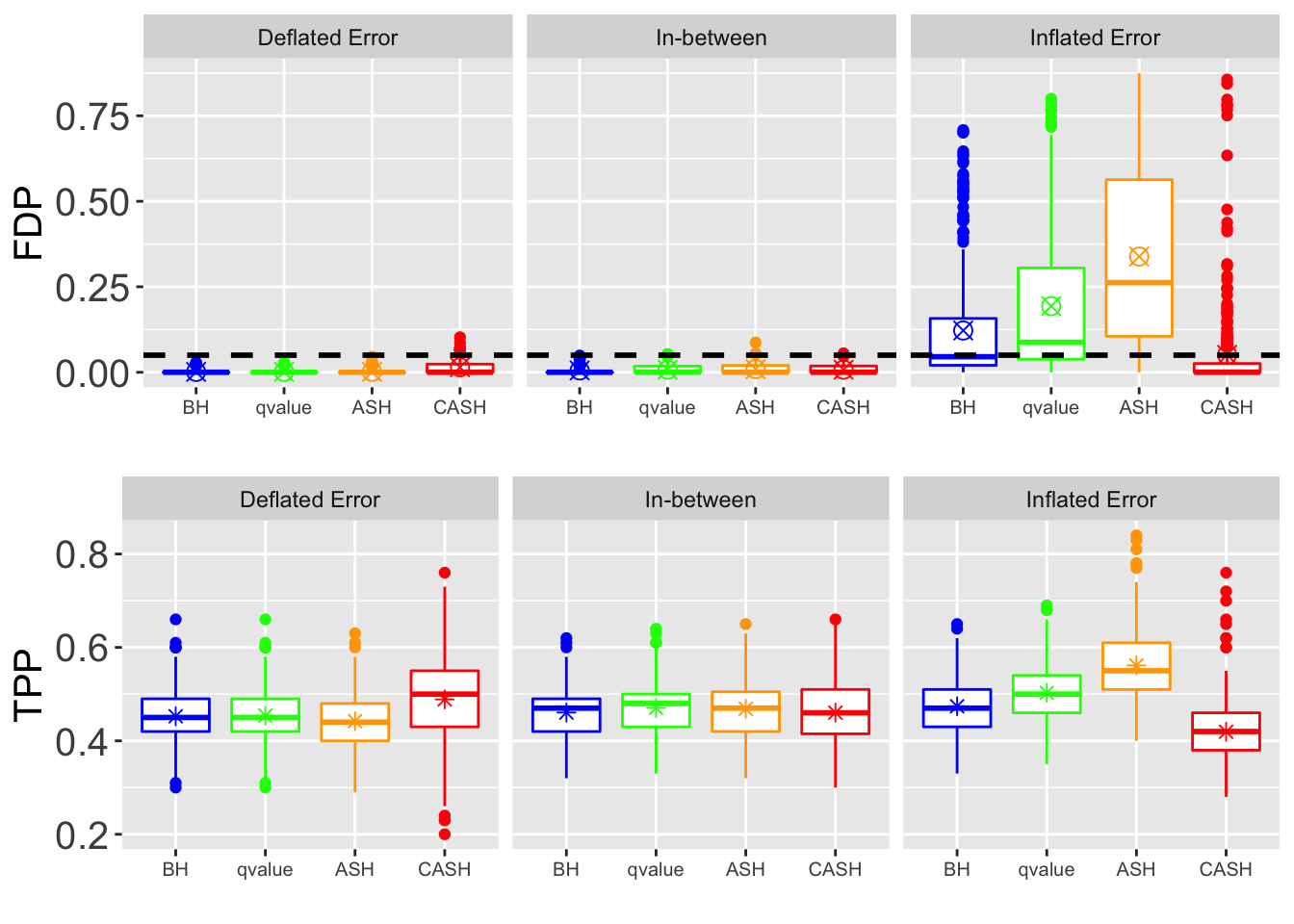
for (j in seq(length(pi0.vec))) {
for (q in c(0.1, 0.05)) {
sd.z <- sapply(z.pi0.list[[j]], sd)
Noise <- cut(sd.z, breaks = c(0, z.under, z.over, Inf), labels = c("Deflated Error", "In-between", "Inflated Error"))
FDR.plot <- ggplot(data = melt(cbind(Noise, as.data.frame(FDP.array[, which(round(q.vec, 4) == q), , j])), id.vars = "Noise"),
aes(x = variable, y = value, col = variable)) +
geom_boxplot() +
stat_summary(fun.y = mean, geom = "point", shape = 8, size = 3) +
# scale_color_manual(values = method.col) +
facet_wrap(~Noise) +
geom_hline(yintercept = q, col = "black", linetype = "dashed", size = 1) +
scale_x_discrete(labels = method.name) +
labs(x = "", y = "FDP") +
theme(legend.position = "none", plot.title = element_text(hjust = 0.5, size = 12.5), axis.title.y = element_text(size = 15), axis.text.x = element_text(size = 7.5), axis.text.y = element_text(size = 15))
# pFDR.plot <- ggplot(data = melt(cbind(Noise, as.data.frame(pFDP.array[, which(q.vec == q), , j])), id.vars = "Noise"),
# aes(x = variable, y = value, col = variable)) +
# geom_boxplot(na.rm = TRUE) +
# stat_summary(fun.y = mean, geom = "point") +
# scale_color_manual(values = method.col) +
# facet_wrap(~Noise) +
# geom_hline(yintercept = q, col = "black", linetype = "dashed", size = 1) +
# scale_x_discrete(labels = method.name) +
# labs(x = "", y = "Positive FDP") +
# theme(legend.position = "none", plot.title = element_text(hjust = 0.5, size = 12.5), axis.title.y = element_text(size = 15), axis.text.x = element_text(size = 7.5), axis.text.y = element_text(size = 15))
power.plot <- ggplot(data = melt(cbind(Noise, as.data.frame(power.array[, which(round(q.vec, 4) == q), , j])), id.vars = "Noise"),
aes(x = variable, y = value, col = variable)) +
geom_boxplot() +
stat_summary(fun.y = mean, geom = "point", shape = 8, size = 3) +
# scale_color_manual(values = method.col) +
facet_wrap(~Noise) +
scale_x_discrete(labels = method.name) +
labs(x = "", y = "TPP") +
theme(legend.position = "none", plot.title = element_text(hjust = 0.5, size = 12.5), axis.title.y = element_text(size = 15), axis.text.x = element_text(size = 7.5), axis.text.y = element_text(size = 15))
grid.arrange(FDR.plot, power.plot
#, top = textGrob(bquote(paste(pi[0] == .(pi0.vec[j]), "; ", sigma[beta] == .(sigma.beta), "; ", .(L), "GDs; ", .(ifelse(w.lambda == 0, "No Constraints", "With Constraints")), "; ", q == .(q))))
)
# pdf(paste("../output/fig/sim_cash_2_", sigma.beta, L, w.lambda, pi0.vec[j], "plot.pdf", sep = "_"), onefile = TRUE)
# grid.arrange(FDR.plot, power.plot, top = textGrob(bquote(paste(pi[0] == .(pi0.vec[j]), "; ", sigma[beta] == .(sigma.beta), "; ", .(L), "GDs; ", .(ifelse(w.lambda == 0, "No Constraints", "With Constraints")), "; ", q == .(q)))))
# dev.off()
}
}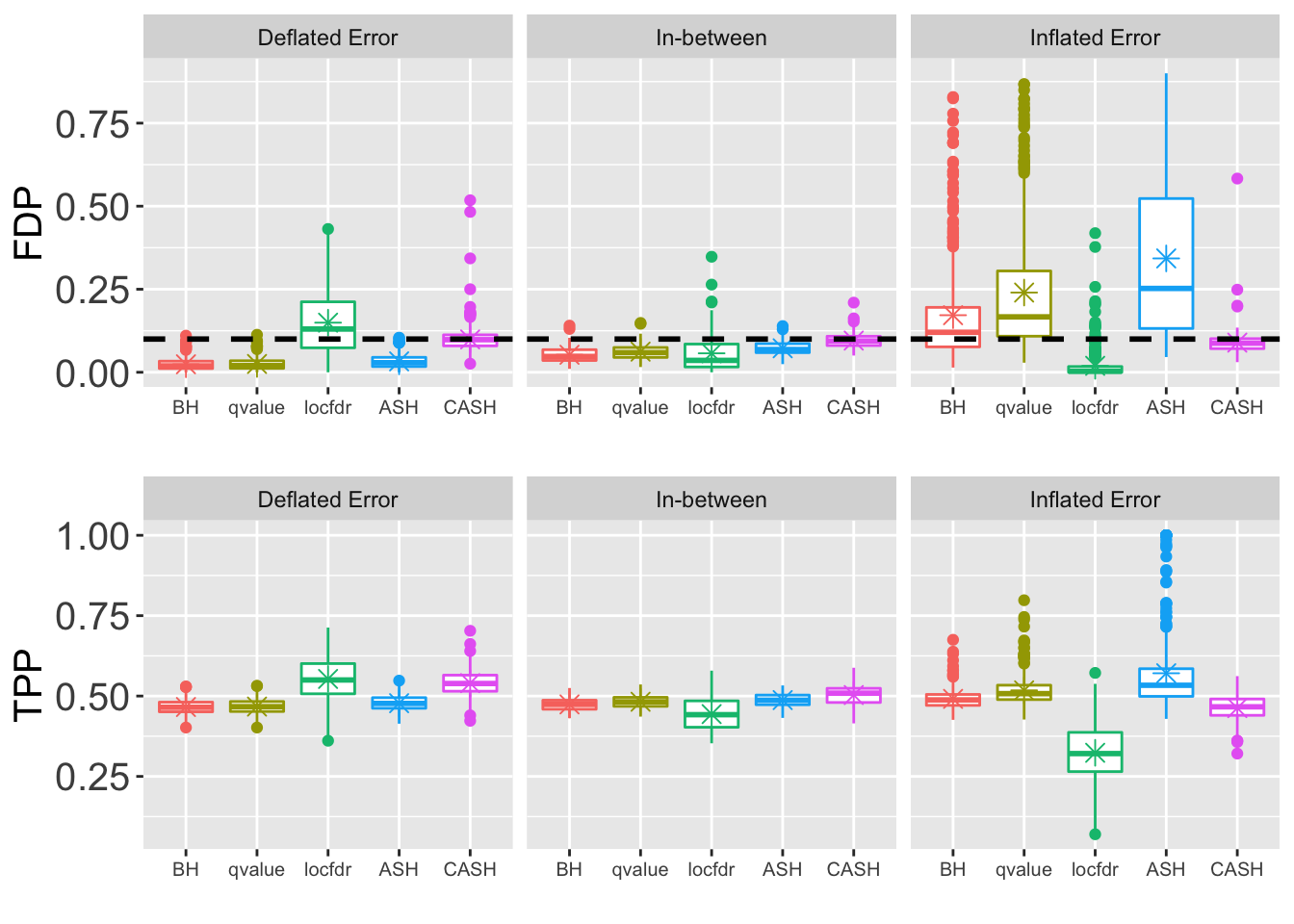
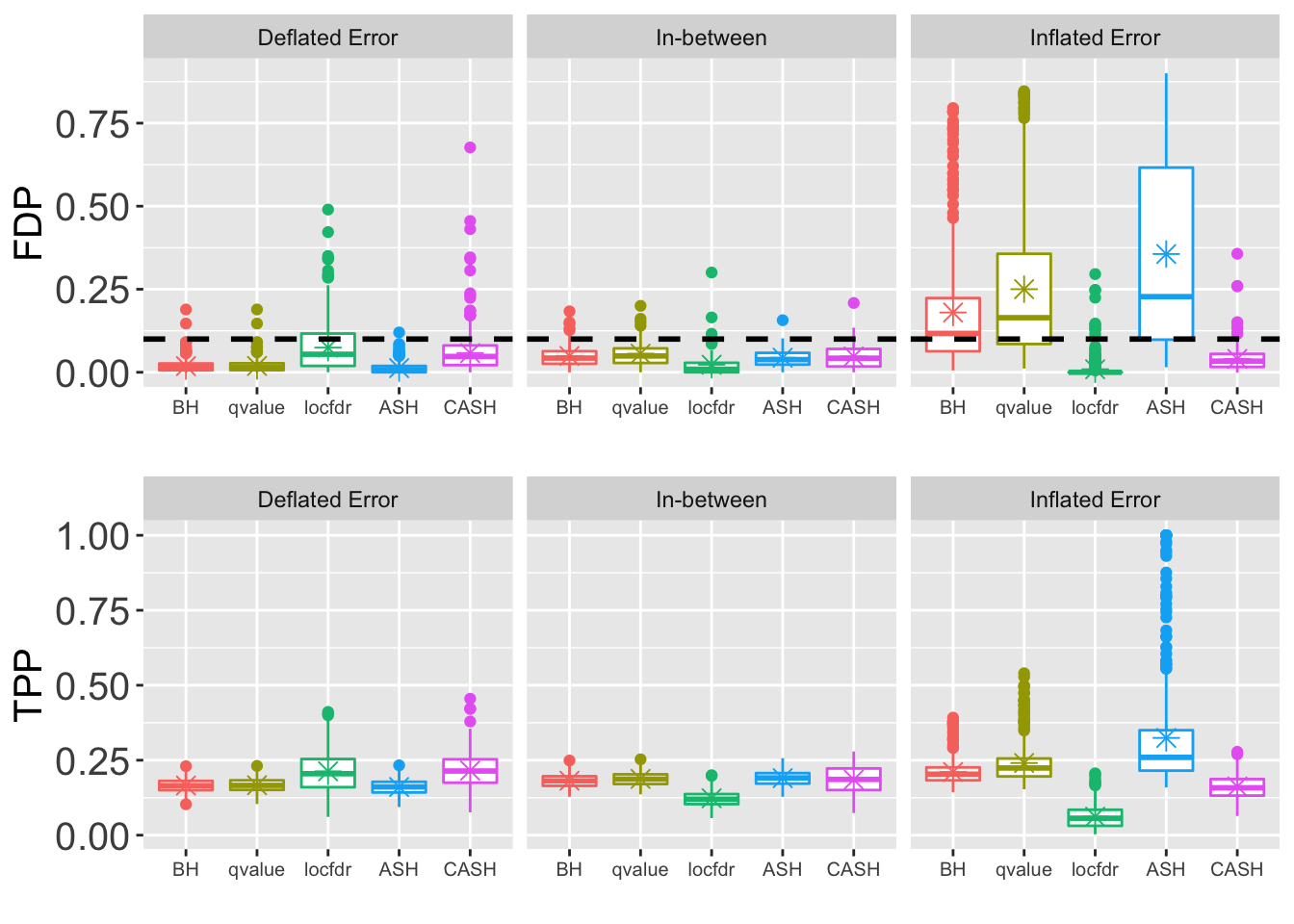
- \(\sigma = 1.5\)
z.over <- 1.025
z.under <- 0.975
method.name <- c("BH", "qvalue", "locfdr", "ASH", "CASH")FDP.array <- pFDP.array <- power.array <- array(0, dim = c(nsim, length(q.vec), length(method.name), length(pi0.vec)))
FDP.summary <- pFDP.summary <- power.summary <- array(0, dim = c(5, length(q.vec), length(method.name), length(pi0.vec)))
for (j in seq(length(pi0.vec))) {
for (k in seq(length(method.name))) {
for (i in seq(nsim)) {
FDP.array[i, , k, j] <- sapply(q.vec, FDP, qvalue = qvalue.pi0.list[[j]][[i]][, k + 1], beta = qvalue.pi0.list[[j]][[i]][, 1])
pFDP.array[i, , k, j] <- sapply(q.vec, pFDP, qvalue = qvalue.pi0.list[[j]][[i]][, k + 1], beta = qvalue.pi0.list[[j]][[i]][, 1])
power.array[i, , k, j] <- sapply(q.vec, power, qvalue = qvalue.pi0.list[[j]][[i]][, k + 1], beta = qvalue.pi0.list[[j]][[i]][, 1])
}
FDP.summary[, , k, j] <- rbind(
avg <- colMeans(FDP.array[, , k, j]),
sd <- apply(FDP.array[, , k, j], 2, sd),
n <- colSums(!is.na(FDP.array[, , k, j])),
q975 <- apply(FDP.array[, , k, j], 2, quantile, probs = 0.975),
q025 <- apply(FDP.array[, , k, j], 2, quantile, probs = 0.025)
)
pFDP.summary[, , k, j] <- rbind(
avg <- colMeans(pFDP.array[, , k, j], na.rm = TRUE),
sd <- apply(pFDP.array[, , k, j], 2, sd, na.rm = TRUE),
n <- colSums(!is.na(pFDP.array[, , k, j])),
q975 <- apply(pFDP.array[, , k, j], 2, quantile, probs = 0.975, na.rm = TRUE),
q025 <- apply(pFDP.array[, , k, j], 2, quantile, probs = 0.025, na.rm = TRUE)
)
power.summary[, , k, j] <- rbind(
avg <- colMeans(power.array[, , k, j]),
sd <- apply(power.array[, , k, j], 2, sd),
n <- colSums(!is.na(power.array[, , k, j])),
q975 <- apply(power.array[, , k, j], 2, quantile, probs = 0.975),
q025 <- apply(power.array[, , k, j], 2, quantile, probs = 0.025)
)
}
}
for (j in seq(length(pi0.vec))) {
for (q in c(0.1)) {
sd.z <- sapply(z.pi0.list[[j]], sd)
Noise <- cut(sd.z, breaks = c(0, z.under, z.over, Inf), labels = c("Deflated Error", "In-between", "Inflated Error"))
FDR.plot <- ggplot(data = melt(cbind(Noise, as.data.frame(FDP.array[, which(round(q.vec, 4) == q), , j])), id.vars = "Noise"),
aes(x = variable, y = value, col = variable)) +
geom_boxplot() +
stat_summary(fun.y = mean, geom = "point", shape = 8, size = 3) +
# scale_color_manual(values = method.col) +
facet_wrap(~Noise) +
geom_hline(yintercept = q, col = "black", linetype = "dashed", size = 1) +
scale_x_discrete(labels = method.name) +
labs(x = "", y = "FDP") +
theme(legend.position = "none", plot.title = element_text(hjust = 0.5, size = 12.5), axis.title.y = element_text(size = 15), axis.text.x = element_text(size = 7.5), axis.text.y = element_text(size = 15))
# pFDR.plot <- ggplot(data = melt(cbind(Noise, as.data.frame(pFDP.array[, which(q.vec == q), , j])), id.vars = "Noise"),
# aes(x = variable, y = value, col = variable)) +
# geom_boxplot(na.rm = TRUE) +
# stat_summary(fun.y = mean, geom = "point") +
# scale_color_manual(values = method.col) +
# facet_wrap(~Noise) +
# geom_hline(yintercept = q, col = "black", linetype = "dashed", size = 1) +
# scale_x_discrete(labels = method.name) +
# labs(x = "", y = "Positive FDP") +
# theme(legend.position = "none", plot.title = element_text(hjust = 0.5, size = 12.5), axis.title.y = element_text(size = 15), axis.text.x = element_text(size = 7.5), axis.text.y = element_text(size = 15))
power.plot <- ggplot(data = melt(cbind(Noise, as.data.frame(power.array[, which(round(q.vec, 4) == q), , j])), id.vars = "Noise"),
aes(x = variable, y = value, col = variable)) +
geom_boxplot() +
stat_summary(fun.y = mean, geom = "point", shape = 8, size = 3) +
# scale_color_manual(values = method.col) +
facet_wrap(~Noise) +
scale_x_discrete(labels = method.name) +
labs(x = "", y = "TPP") +
theme(legend.position = "none", plot.title = element_text(hjust = 0.5, size = 12.5), axis.title.y = element_text(size = 15), axis.text.x = element_text(size = 7.5), axis.text.y = element_text(size = 15))
grid.arrange(FDR.plot, power.plot
#, top = textGrob(bquote(paste(pi[0] == .(pi0.vec[j]), "; ", sigma[beta] == .(sigma.beta), "; ", .(L), "GDs; ", .(ifelse(w.lambda == 0, "No Constraints", "With Constraints")), "; ", q == .(q))))
)
# pdf(paste("../output/fig/sim_cash_2_", sigma.beta, L, w.lambda, pi0.vec[j], "plot.pdf", sep = "_"), onefile = TRUE)
# grid.arrange(FDR.plot, power.plot, top = textGrob(bquote(paste(pi[0] == .(pi0.vec[j]), "; ", sigma[beta] == .(sigma.beta), "; ", .(L), "GDs; ", .(ifelse(w.lambda == 0, "No Constraints", "With Constraints")), "; ", q == .(q)))))
# dev.off()
}
}
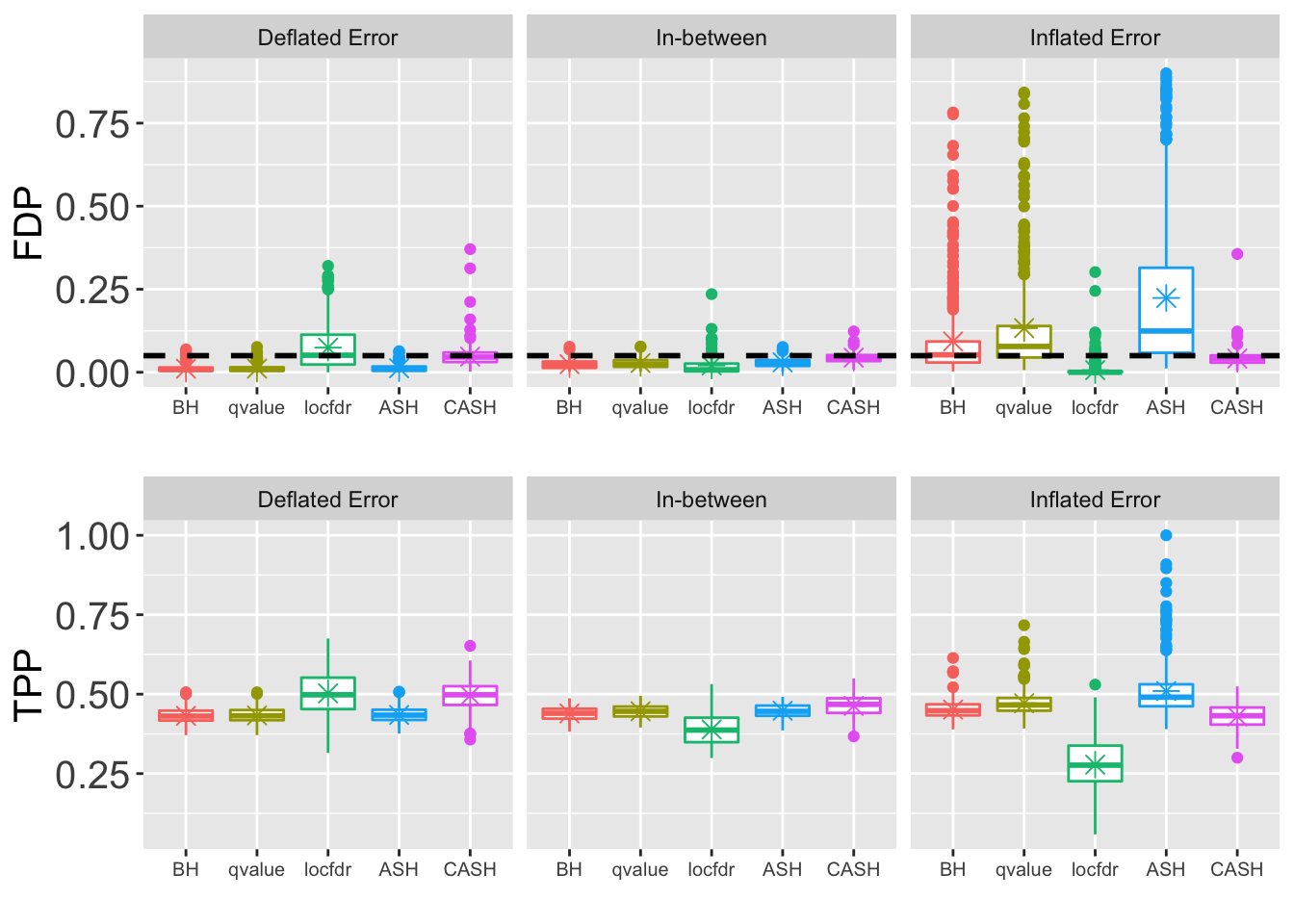
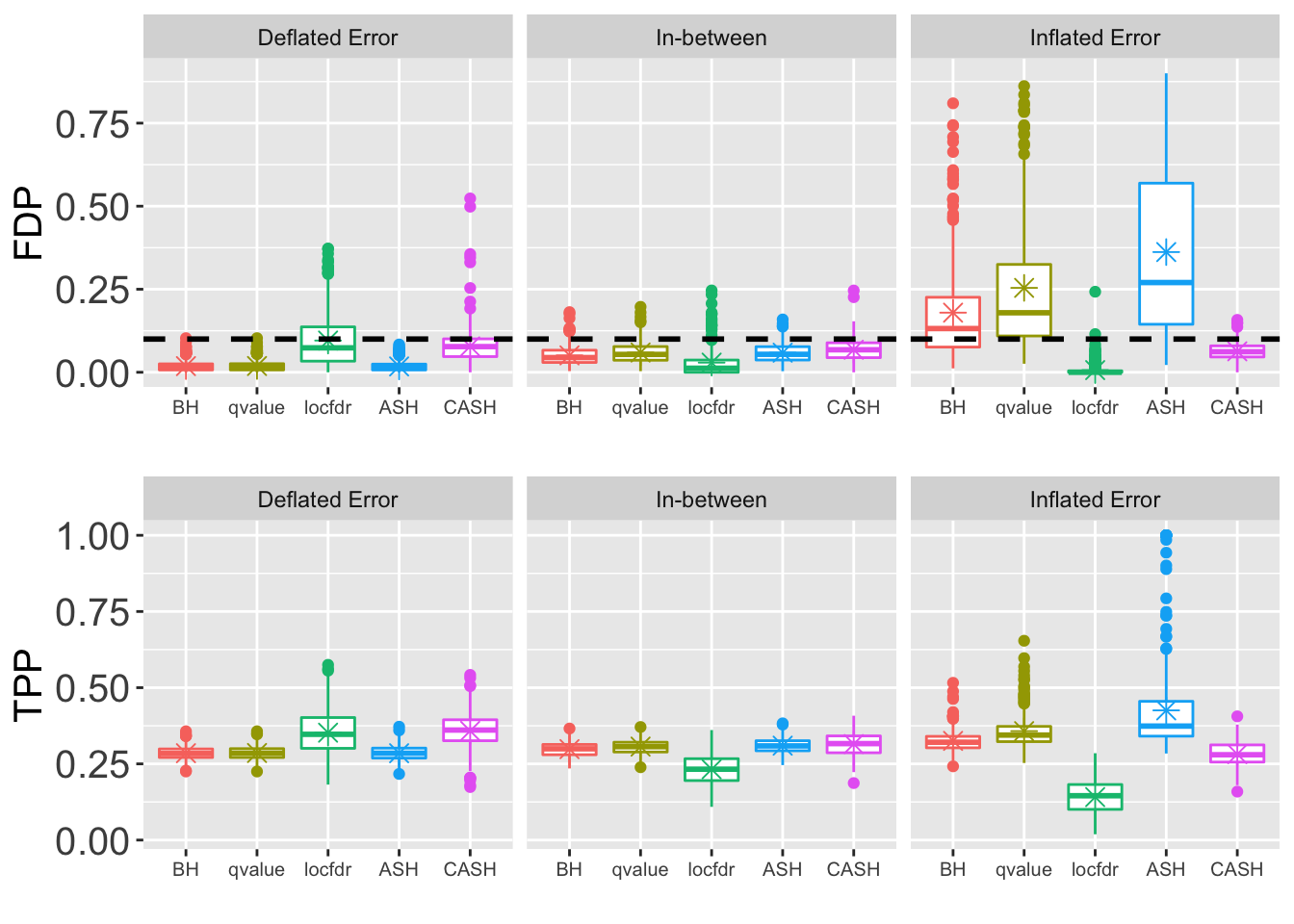
- \(\sigma = 2\)
z.over <- 1.05
z.under <- 0.95
method.name <- c("BH", "qvalue", "locfdr", "ASH", "CASH")FDP.array <- pFDP.array <- power.array <- array(0, dim = c(nsim, length(q.vec), length(method.name), length(pi0.vec)))
FDP.summary <- pFDP.summary <- power.summary <- array(0, dim = c(5, length(q.vec), length(method.name), length(pi0.vec)))
for (j in seq(length(pi0.vec))) {
for (k in seq(length(method.name))) {
for (i in seq(nsim)) {
FDP.array[i, , k, j] <- sapply(q.vec, FDP, qvalue = qvalue.pi0.list[[j]][[i]][, k + 1], beta = qvalue.pi0.list[[j]][[i]][, 1])
pFDP.array[i, , k, j] <- sapply(q.vec, pFDP, qvalue = qvalue.pi0.list[[j]][[i]][, k + 1], beta = qvalue.pi0.list[[j]][[i]][, 1])
power.array[i, , k, j] <- sapply(q.vec, power, qvalue = qvalue.pi0.list[[j]][[i]][, k + 1], beta = qvalue.pi0.list[[j]][[i]][, 1])
}
FDP.summary[, , k, j] <- rbind(
avg <- colMeans(FDP.array[, , k, j]),
sd <- apply(FDP.array[, , k, j], 2, sd),
n <- colSums(!is.na(FDP.array[, , k, j])),
q975 <- apply(FDP.array[, , k, j], 2, quantile, probs = 0.975),
q025 <- apply(FDP.array[, , k, j], 2, quantile, probs = 0.025)
)
pFDP.summary[, , k, j] <- rbind(
avg <- colMeans(pFDP.array[, , k, j], na.rm = TRUE),
sd <- apply(pFDP.array[, , k, j], 2, sd, na.rm = TRUE),
n <- colSums(!is.na(pFDP.array[, , k, j])),
q975 <- apply(pFDP.array[, , k, j], 2, quantile, probs = 0.975, na.rm = TRUE),
q025 <- apply(pFDP.array[, , k, j], 2, quantile, probs = 0.025, na.rm = TRUE)
)
power.summary[, , k, j] <- rbind(
avg <- colMeans(power.array[, , k, j]),
sd <- apply(power.array[, , k, j], 2, sd),
n <- colSums(!is.na(power.array[, , k, j])),
q975 <- apply(power.array[, , k, j], 2, quantile, probs = 0.975),
q025 <- apply(power.array[, , k, j], 2, quantile, probs = 0.025)
)
}
}
for (j in seq(length(pi0.vec))) {
for (q in c(0.1)) {
sd.z <- sapply(z.pi0.list[[j]], sd)
Noise <- cut(sd.z, breaks = c(0, z.under, z.over, Inf), labels = c("Deflated Error", "In-between", "Inflated Error"))
FDR.plot <- ggplot(data = melt(cbind(Noise, as.data.frame(FDP.array[, which(round(q.vec, 4) == q), , j])), id.vars = "Noise"),
aes(x = variable, y = value, col = variable)) +
geom_boxplot() +
stat_summary(fun.y = mean, geom = "point", shape = 8, size = 3) +
# scale_color_manual(values = method.col) +
facet_wrap(~Noise) +
geom_hline(yintercept = q, col = "black", linetype = "dashed", size = 1) +
scale_x_discrete(labels = method.name) +
labs(x = "", y = "FDP") +
theme(legend.position = "none", plot.title = element_text(hjust = 0.5, size = 12.5), axis.title.y = element_text(size = 15), axis.text.x = element_text(size = 7.5), axis.text.y = element_text(size = 15))
# pFDR.plot <- ggplot(data = melt(cbind(Noise, as.data.frame(pFDP.array[, which(q.vec == q), , j])), id.vars = "Noise"),
# aes(x = variable, y = value, col = variable)) +
# geom_boxplot(na.rm = TRUE) +
# stat_summary(fun.y = mean, geom = "point") +
# scale_color_manual(values = method.col) +
# facet_wrap(~Noise) +
# geom_hline(yintercept = q, col = "black", linetype = "dashed", size = 1) +
# scale_x_discrete(labels = method.name) +
# labs(x = "", y = "Positive FDP") +
# theme(legend.position = "none", plot.title = element_text(hjust = 0.5, size = 12.5), axis.title.y = element_text(size = 15), axis.text.x = element_text(size = 7.5), axis.text.y = element_text(size = 15))
power.plot <- ggplot(data = melt(cbind(Noise, as.data.frame(power.array[, which(round(q.vec, 4) == q), , j])), id.vars = "Noise"),
aes(x = variable, y = value, col = variable)) +
geom_boxplot() +
stat_summary(fun.y = mean, geom = "point", shape = 8, size = 3) +
# scale_color_manual(values = method.col) +
facet_wrap(~Noise) +
scale_x_discrete(labels = method.name) +
labs(x = "", y = "TPP") +
theme(legend.position = "none", plot.title = element_text(hjust = 0.5, size = 12.5), axis.title.y = element_text(size = 15), axis.text.x = element_text(size = 7.5), axis.text.y = element_text(size = 15))
grid.arrange(FDR.plot, power.plot
#, top = textGrob(bquote(paste(pi[0] == .(pi0.vec[j]), "; ", sigma[beta] == .(sigma.beta), "; ", .(L), "GDs; ", .(ifelse(w.lambda == 0, "No Constraints", "With Constraints")), "; ", q == .(q))))
)
# pdf(paste("../output/fig/sim_cash_2_", sigma.beta, L, w.lambda, pi0.vec[j], "plot.pdf", sep = "_"), onefile = TRUE)
# grid.arrange(FDR.plot, power.plot, top = textGrob(bquote(paste(pi[0] == .(pi0.vec[j]), "; ", sigma[beta] == .(sigma.beta), "; ", .(L), "GDs; ", .(ifelse(w.lambda == 0, "No Constraints", "With Constraints")), "; ", q == .(q)))))
# dev.off()
}
}
Session information
sessionInfo()R version 3.4.3 (2017-11-30)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS High Sierra 10.13.4
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] grid stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] gridExtra_2.3 ggplot2_2.2.1 reshape2_1.4.3
[4] locfdr_1.1-8 ashr_2.2-2 Rmosek_8.0.69
[7] PolynomF_1.0-1 CVXR_0.95 REBayes_1.2
[10] Matrix_1.2-12 SQUAREM_2017.10-1 EQL_1.0-0
[13] ttutils_1.0-1
loaded via a namespace (and not attached):
[1] gmp_0.5-13.1 Rcpp_0.12.16 pillar_1.0.1
[4] plyr_1.8.4 compiler_3.4.3 git2r_0.21.0
[7] R.methodsS3_1.7.1 R.utils_2.6.0 iterators_1.0.9
[10] tools_3.4.3 digest_0.6.15 bit_1.1-12
[13] tibble_1.4.1 gtable_0.2.0 evaluate_0.10.1
[16] lattice_0.20-35 rlang_0.1.6 foreach_1.4.4
[19] yaml_2.1.18 parallel_3.4.3 Rmpfr_0.6-1
[22] ECOSolveR_0.4 stringr_1.3.0 knitr_1.20
[25] rprojroot_1.3-2 bit64_0.9-7 R6_2.2.2
[28] rmarkdown_1.9 magrittr_1.5 scales_0.5.0
[31] splines_3.4.3 MASS_7.3-47 backports_1.1.2
[34] codetools_0.2-15 htmltools_0.3.6 scs_1.1-1
[37] colorspace_1.3-2 labeling_0.3 stringi_1.1.6
[40] lazyeval_0.2.1 munsell_0.4.3 doParallel_1.0.11
[43] pscl_1.5.2 truncnorm_1.0-7 R.oo_1.21.0 This R Markdown site was created with workflowr