Simulation with Realistic Standard Error
Lei Sun
2017-05-27
Last updated: 2017-06-01
Code version: a40ffff
library(ashr)
library(edgeR)
library(limma)
library(pROC)source("../code/gdash.R")Read in data
data = readRDS("../data/liver.rds")
ngene = 1e4
data = top_gene_selection(ngene, data)$dataGlobal Null
\[ g \equiv 0 \ . \]
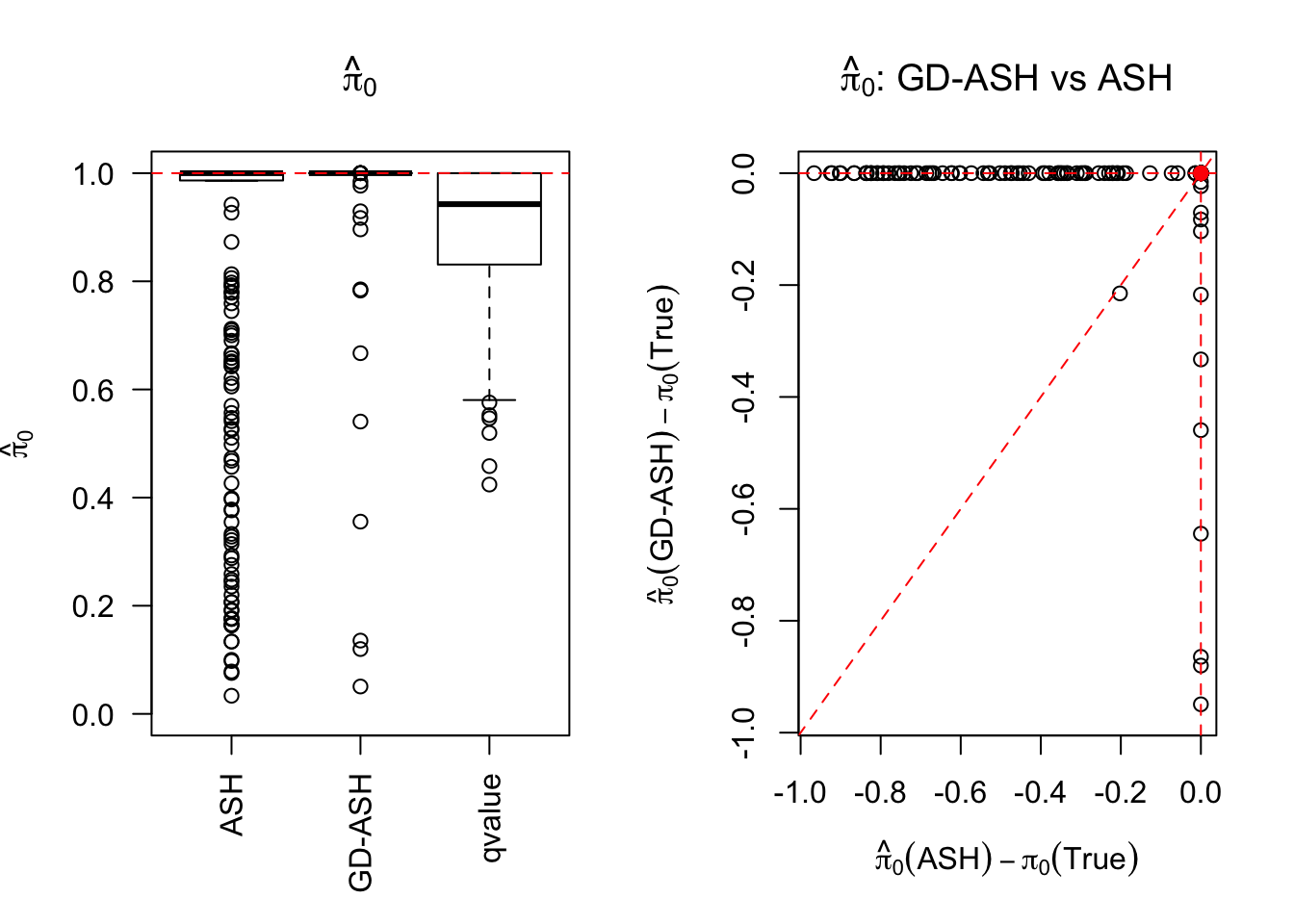
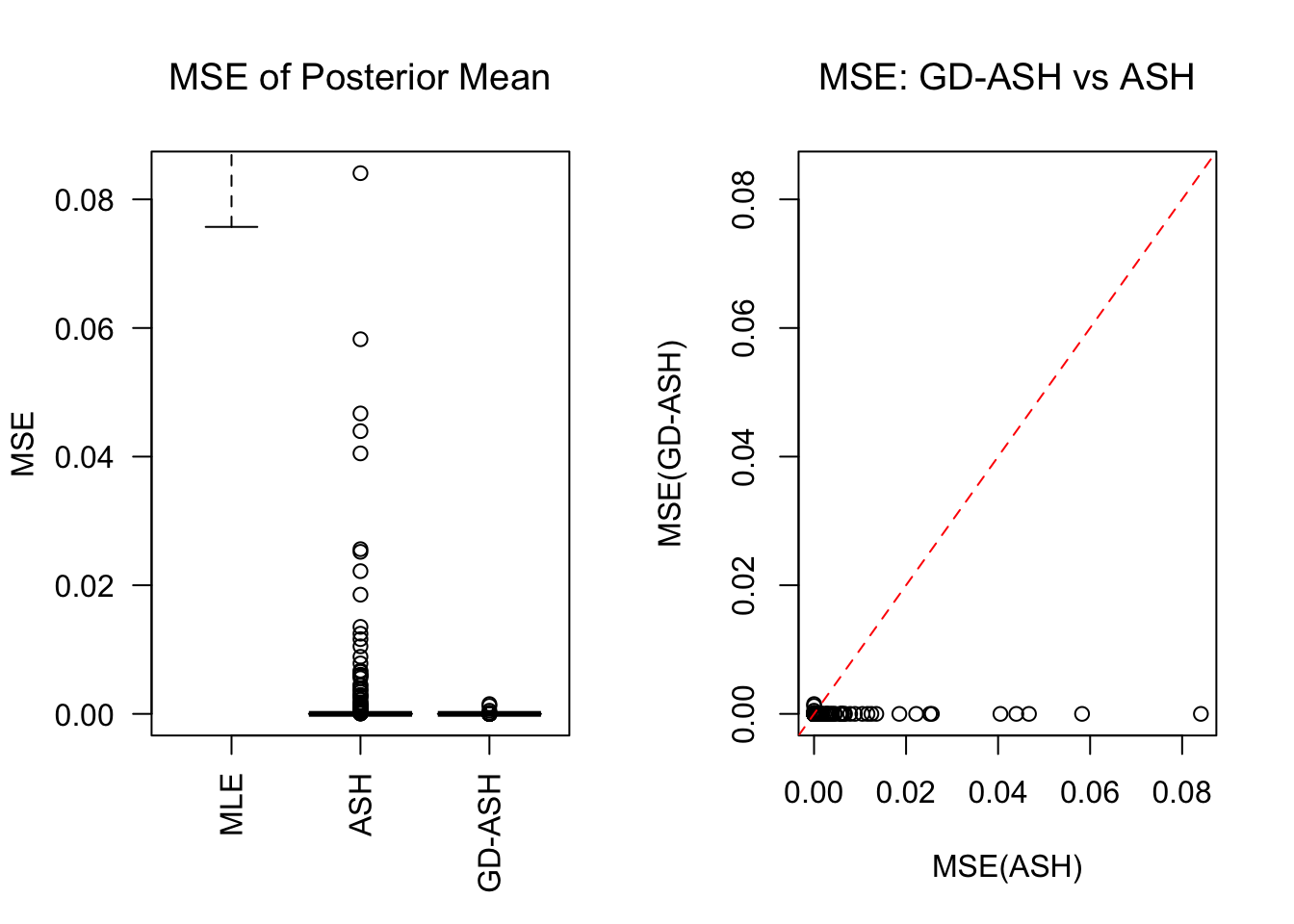
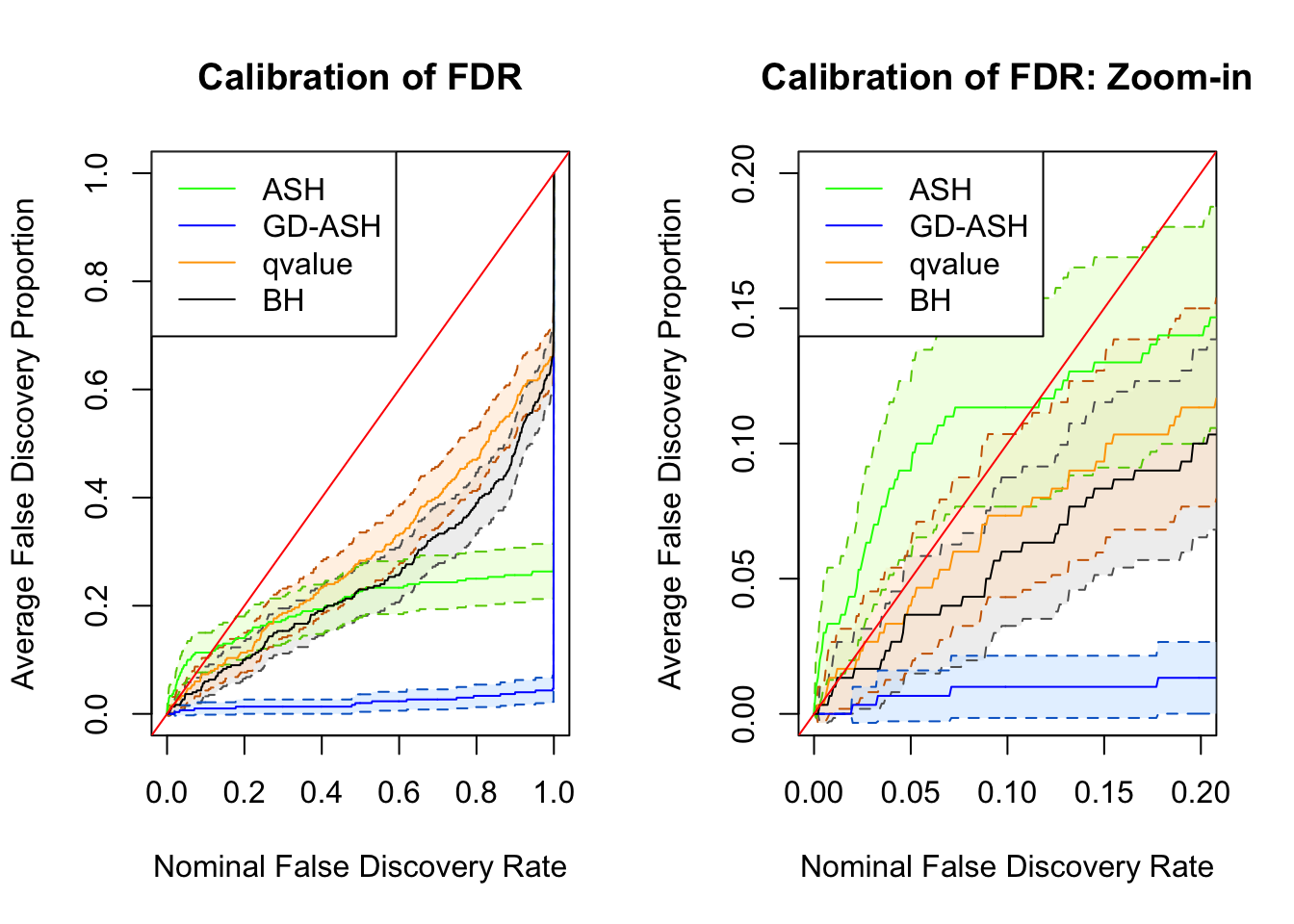
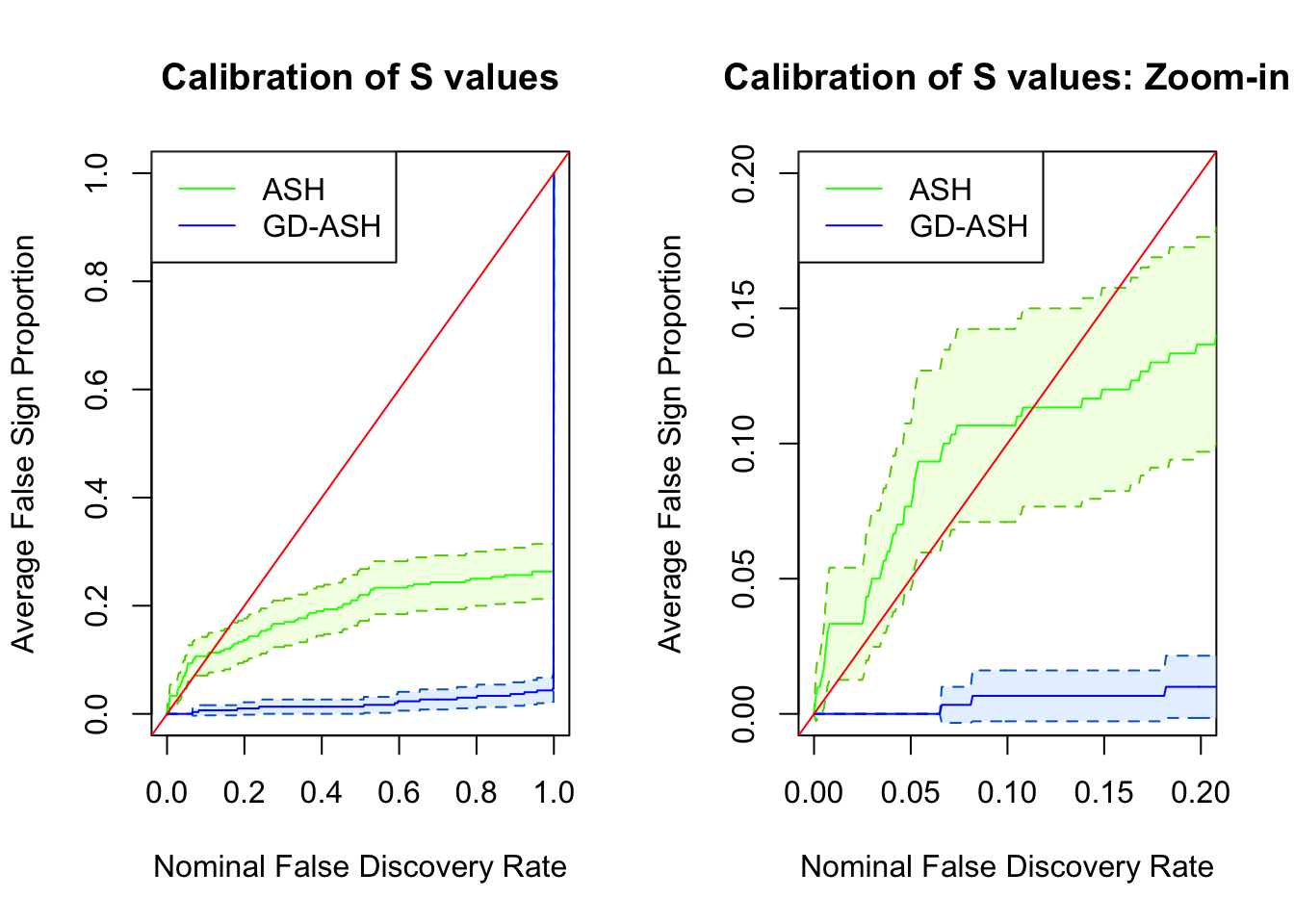
SNR = 0
\[ g = 0.6\delta_0 + 0.3N\left(0, \sigma^2\right) + 0.1N\left(0, \left(2.65\sigma\right)^2\right) \ . \]
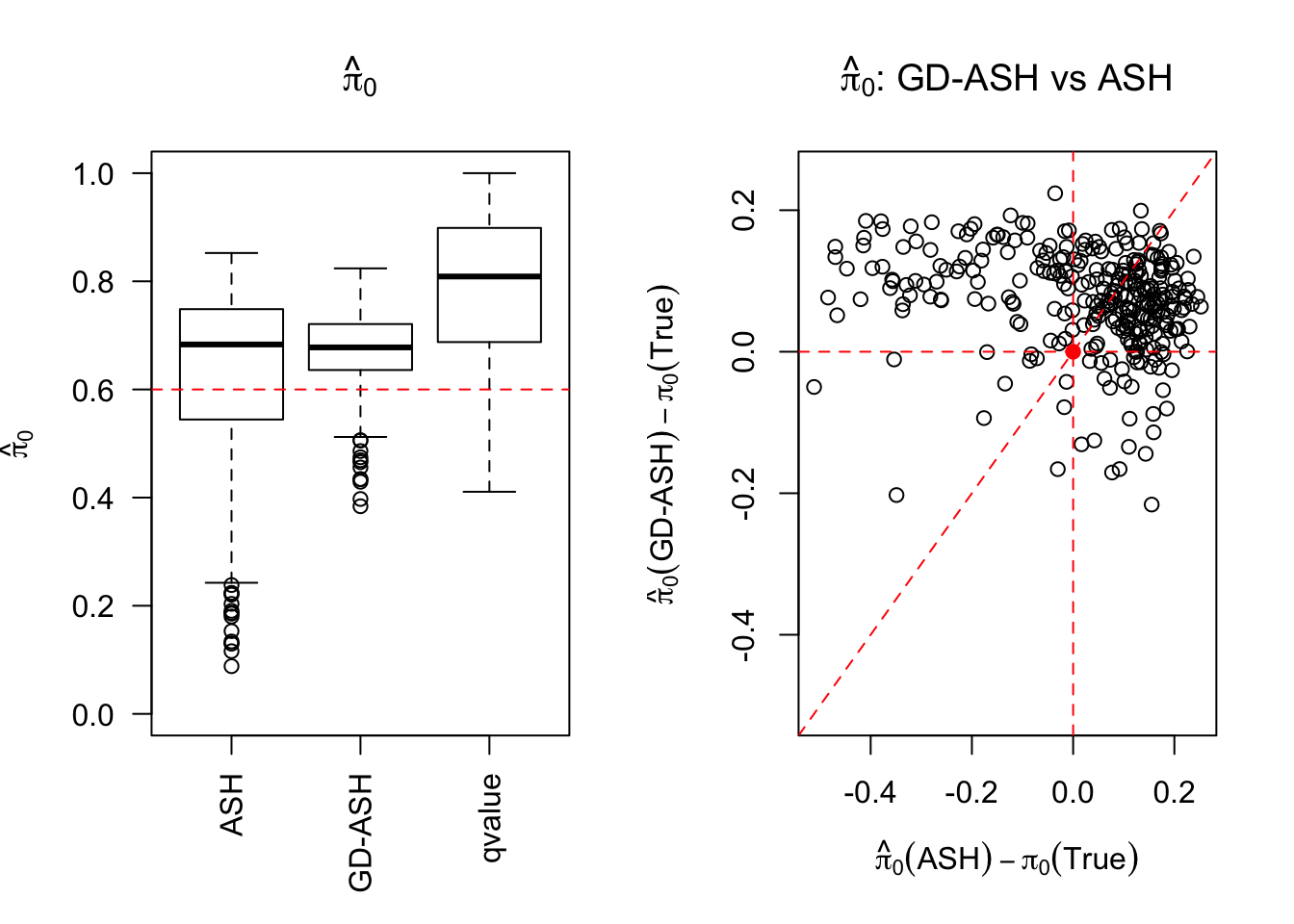
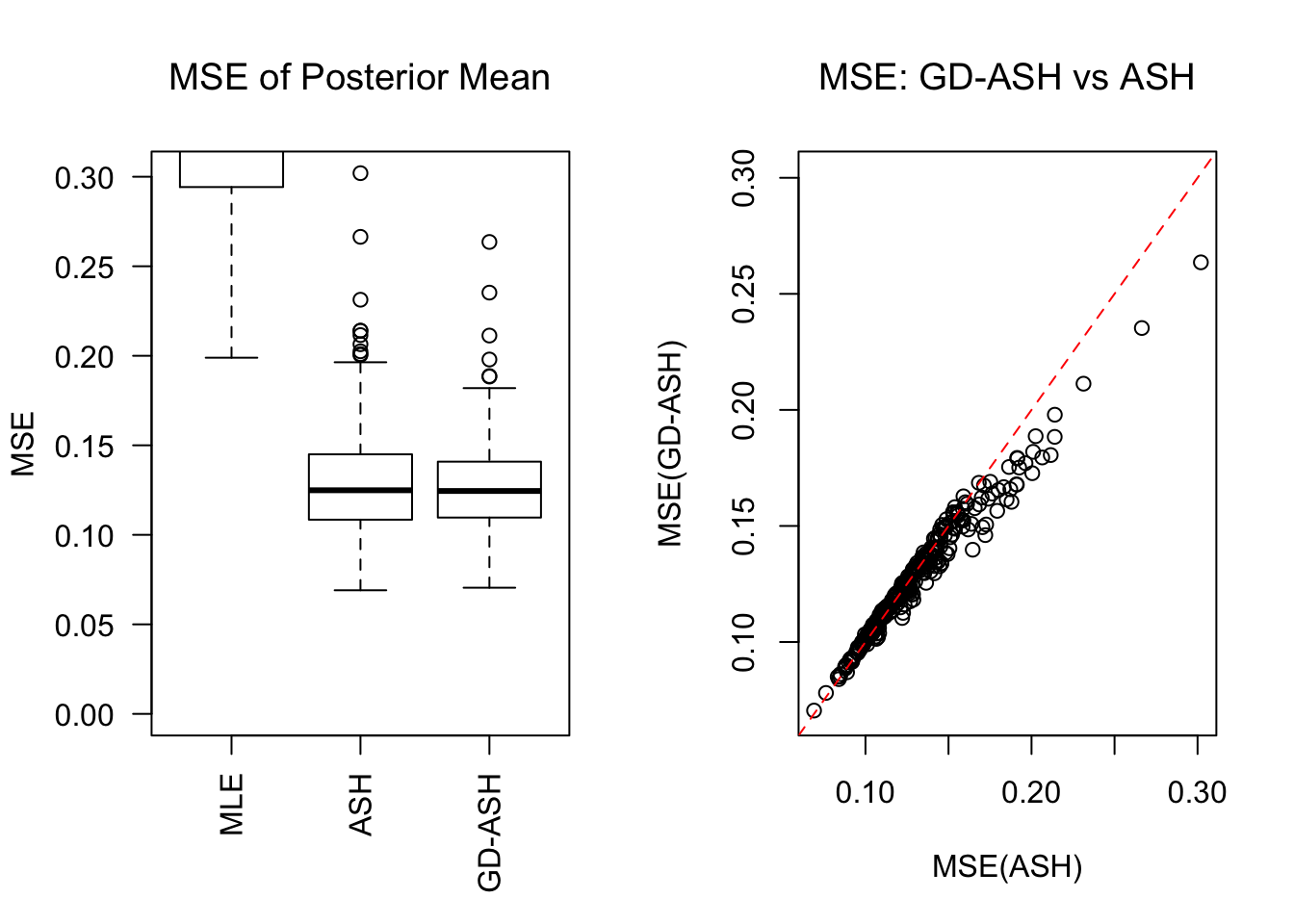
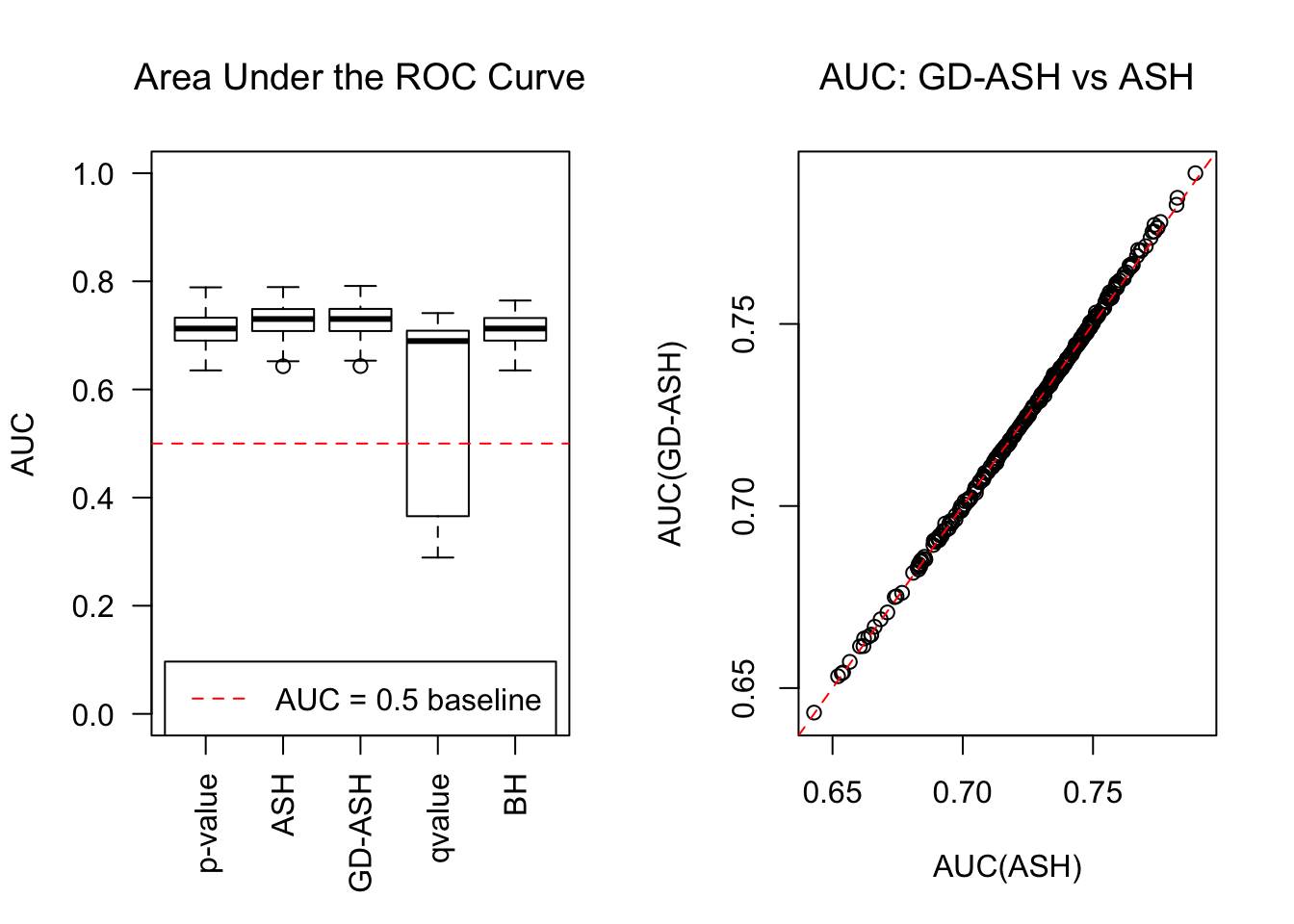
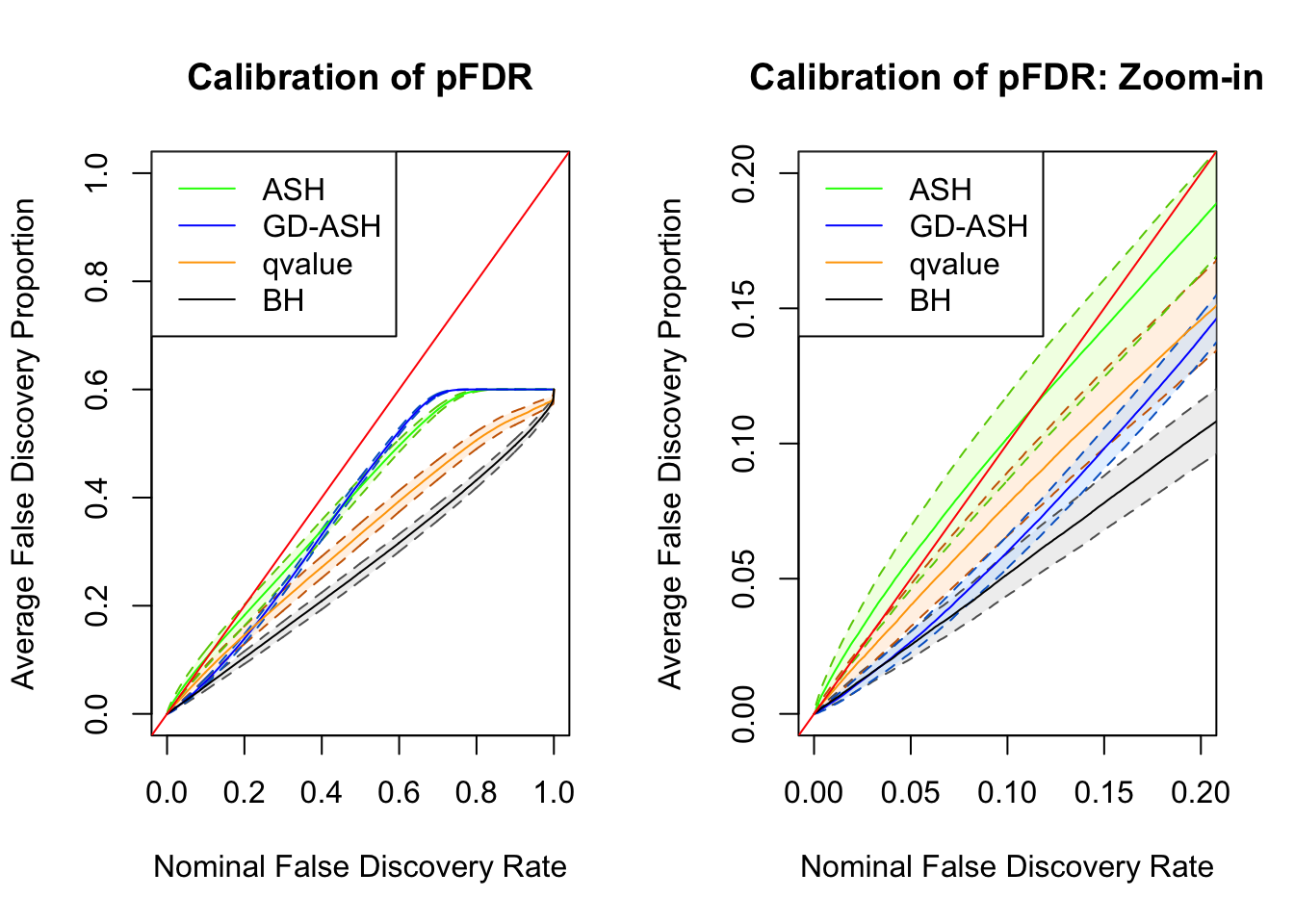
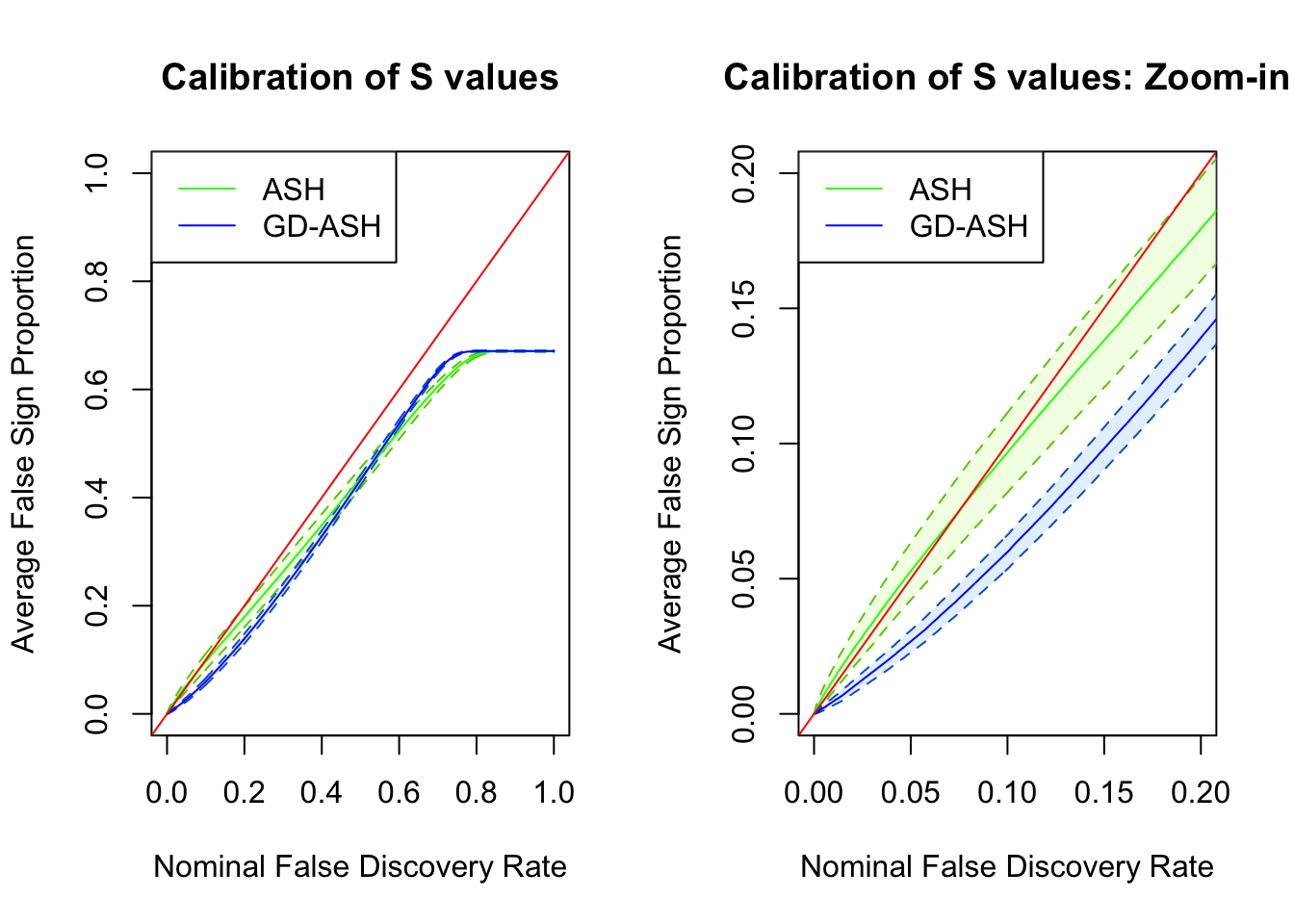
SNR = 2
\[ g = 0.6\delta_0 + 0.3N\left(0, \sigma^2\right) + 0.1N\left(0, \left(3.58\sigma\right)^2\right) \ . \]
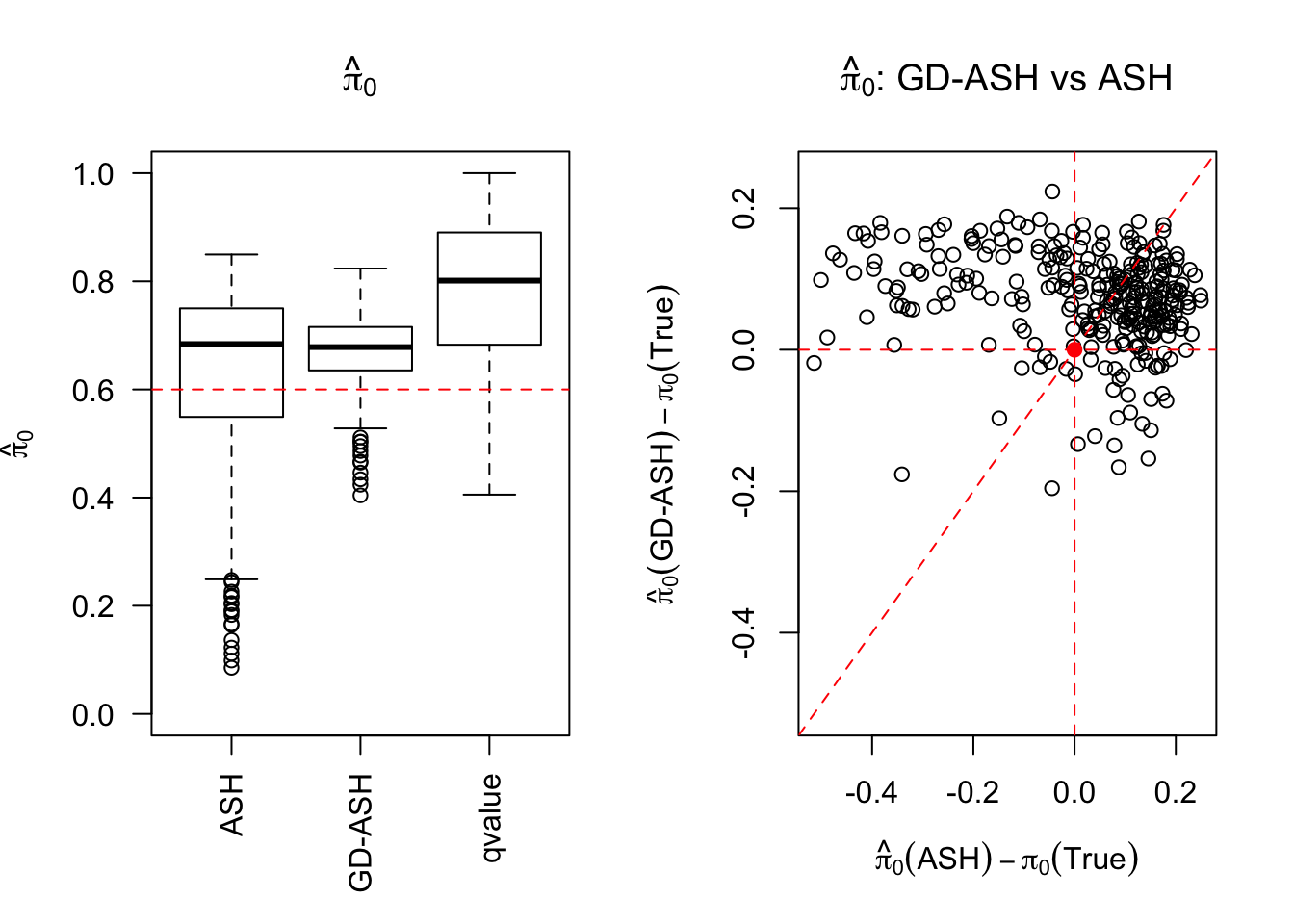
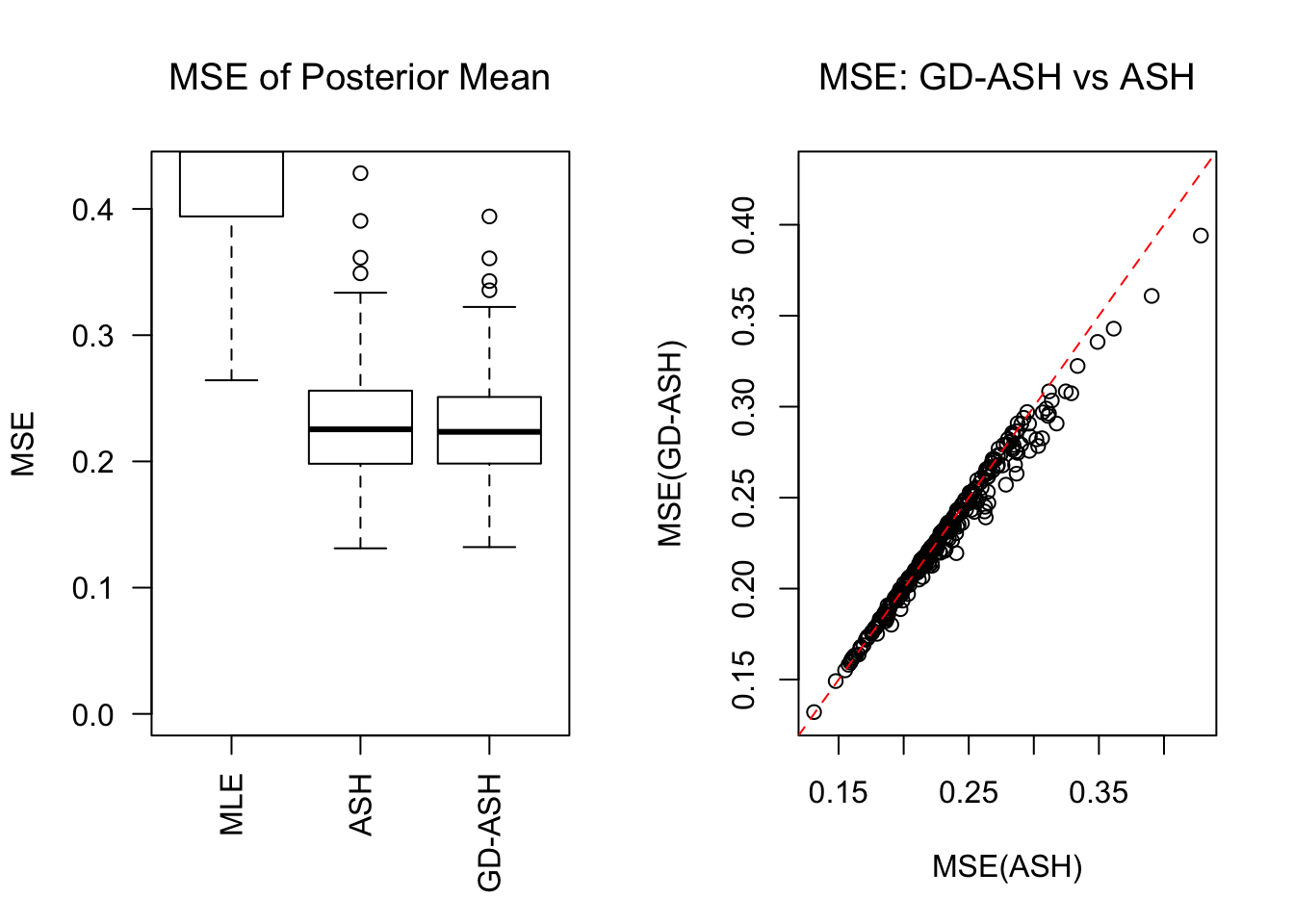
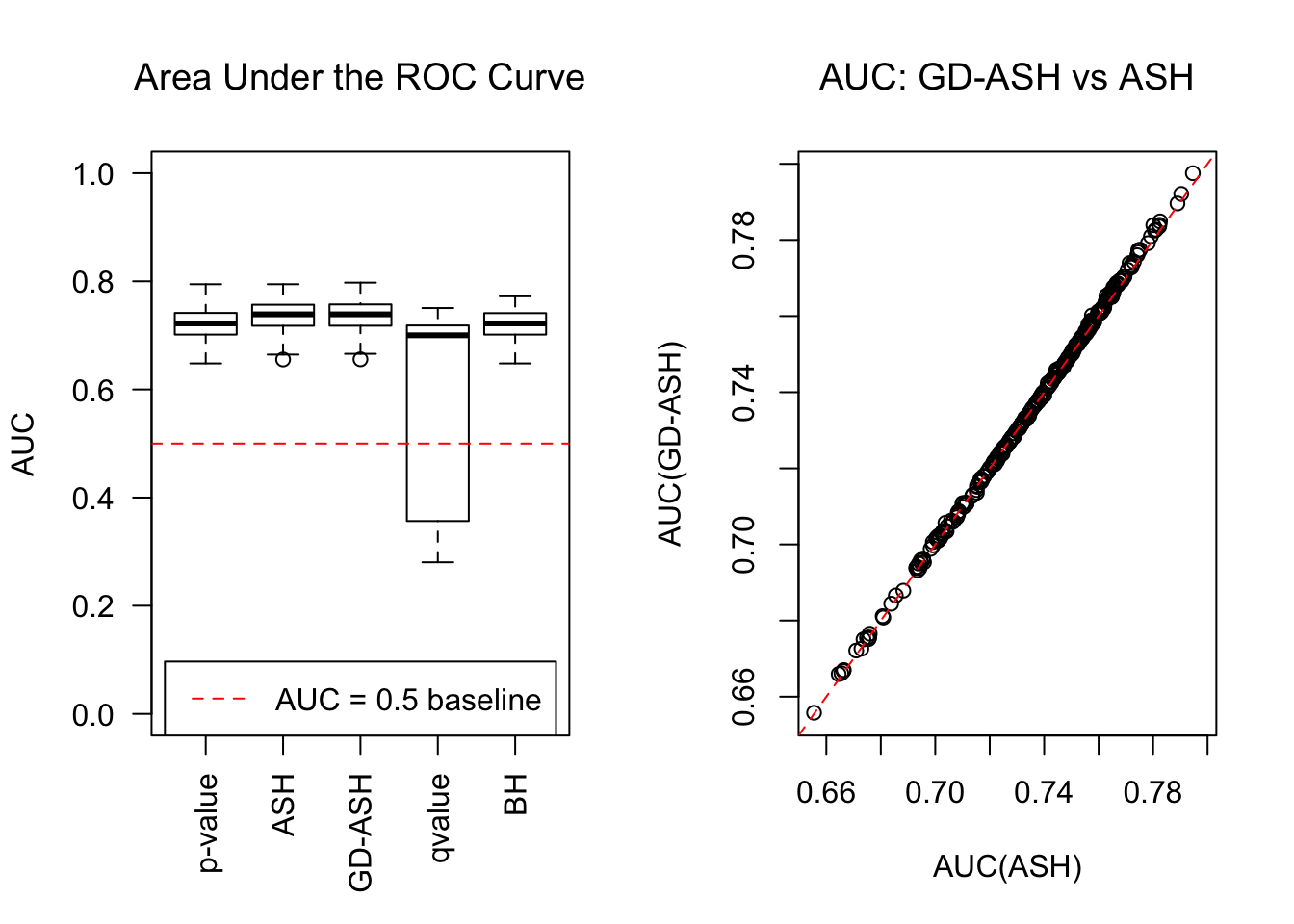
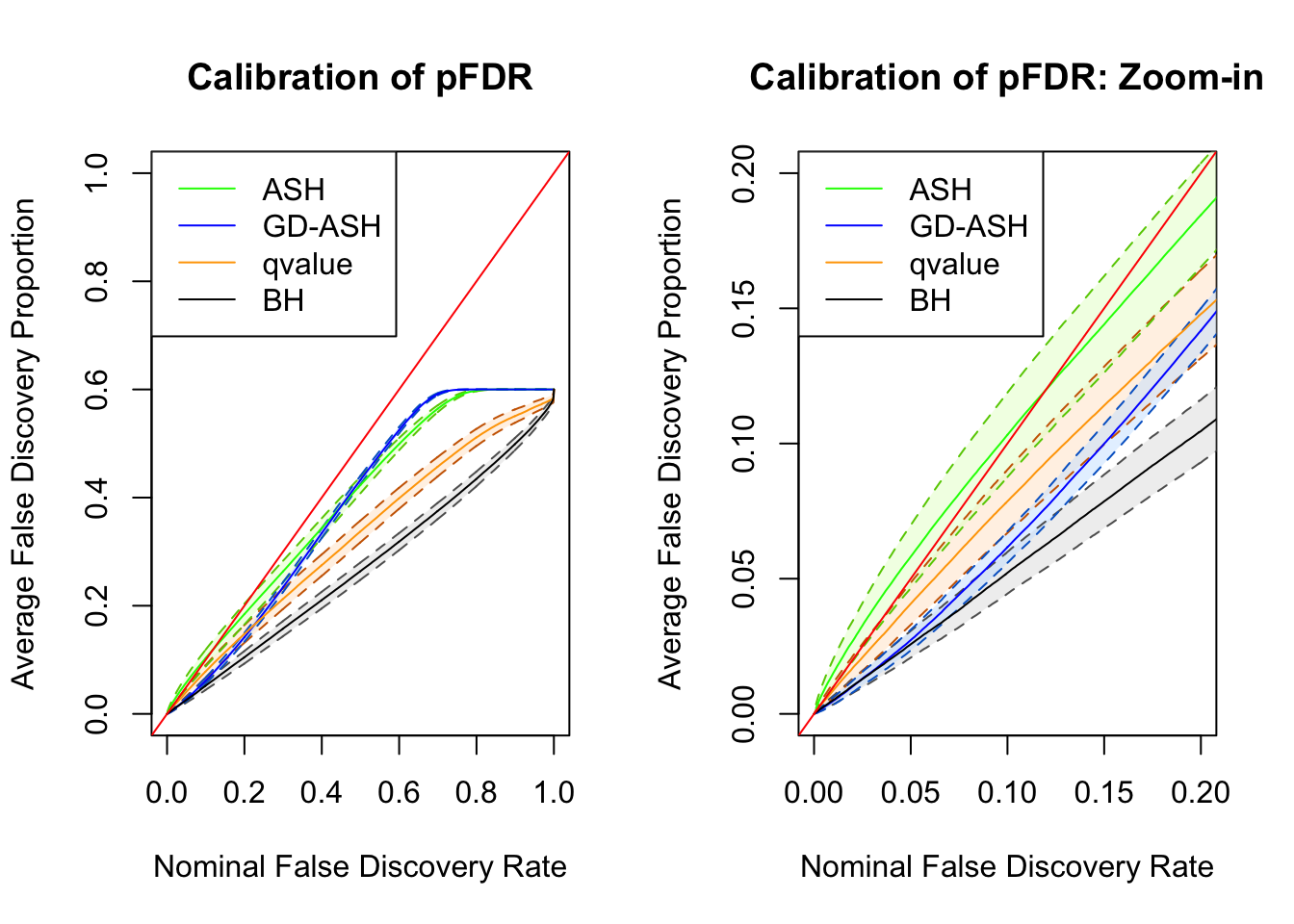
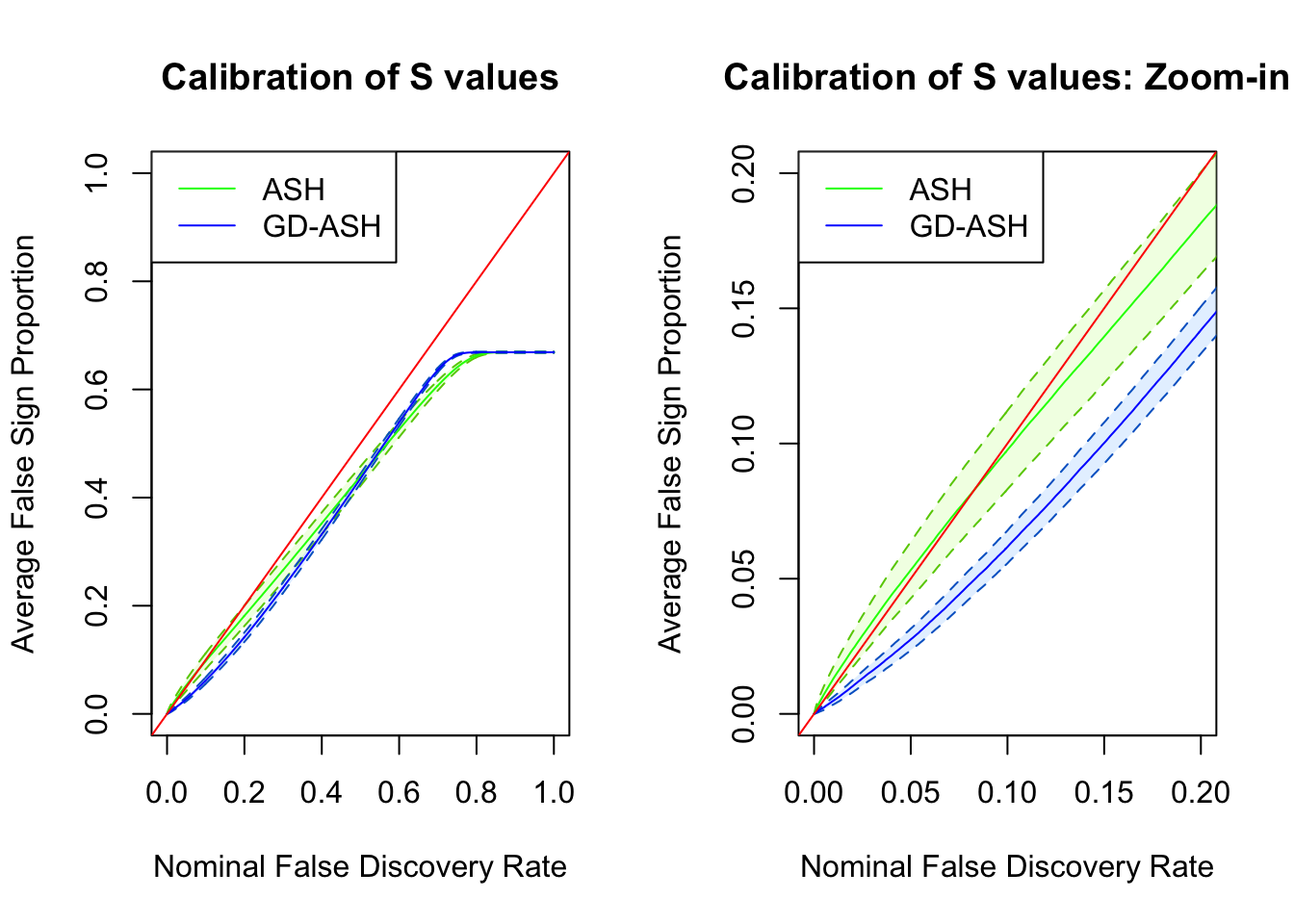
SNR = 5
\[ g = 0.6\delta_0 + 0.3N\left(0, \left(2\sigma\right)^2\right) + 0.1N\left(0, \left(4.43\sigma\right)^2\right) \ . \]
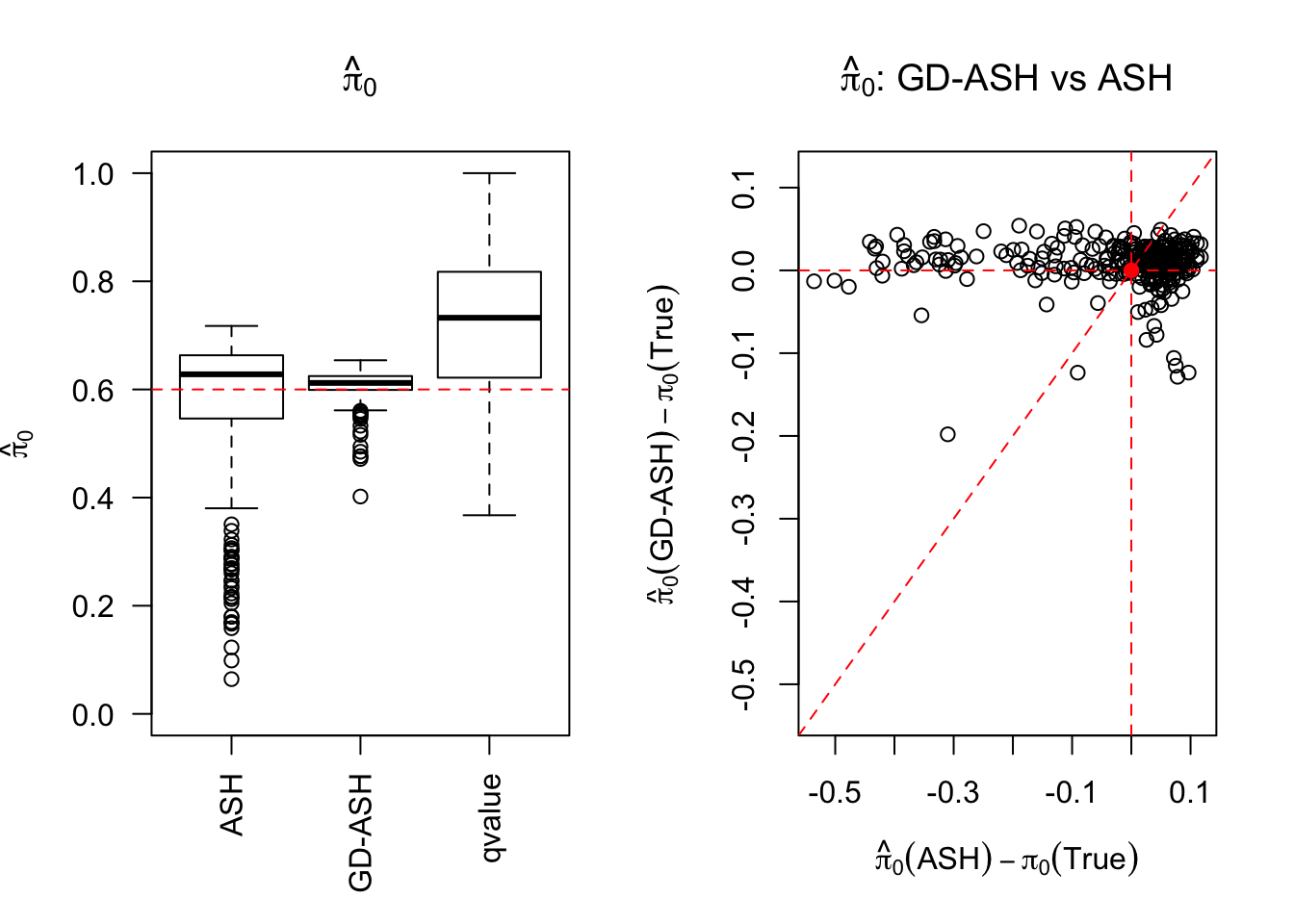
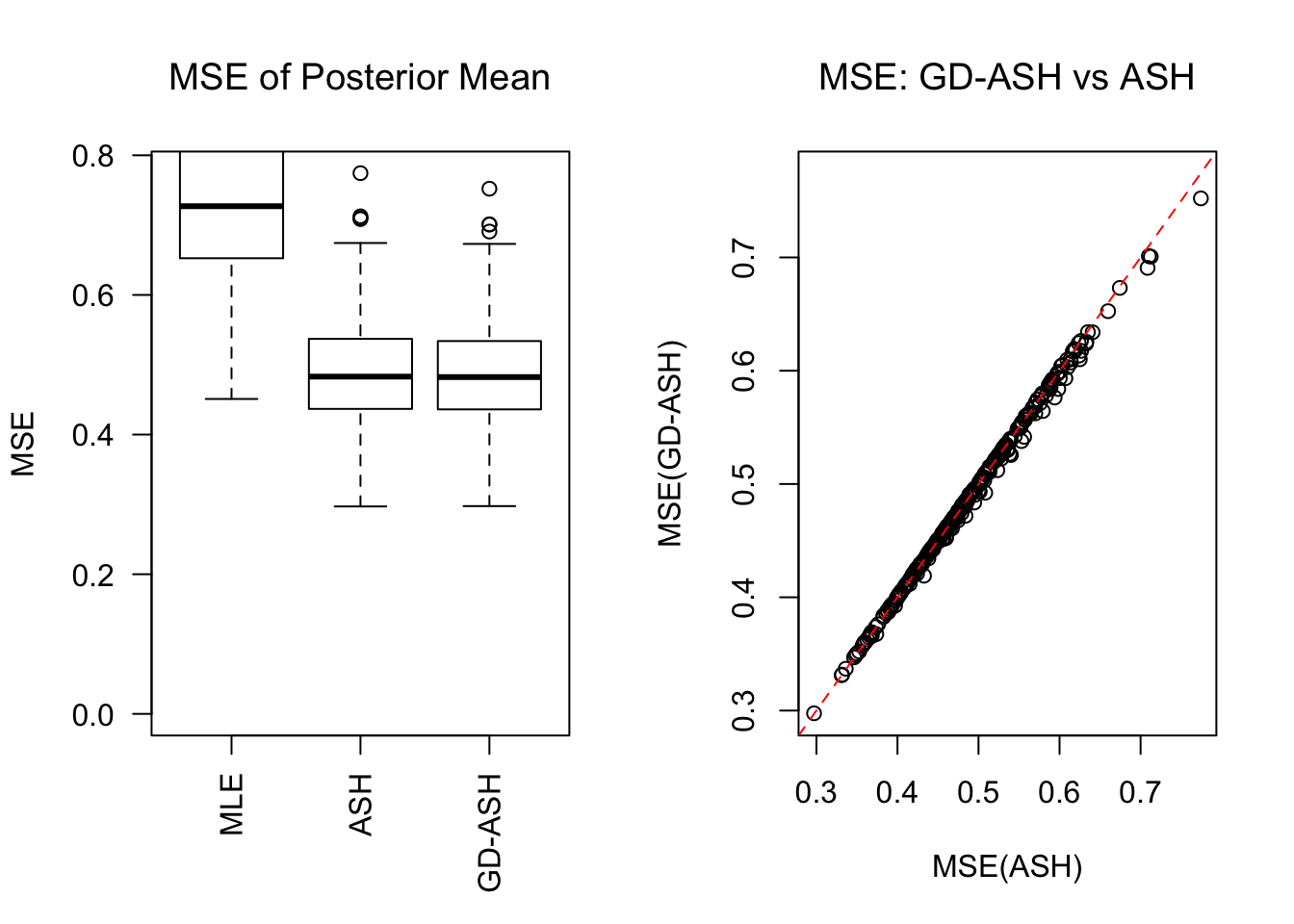
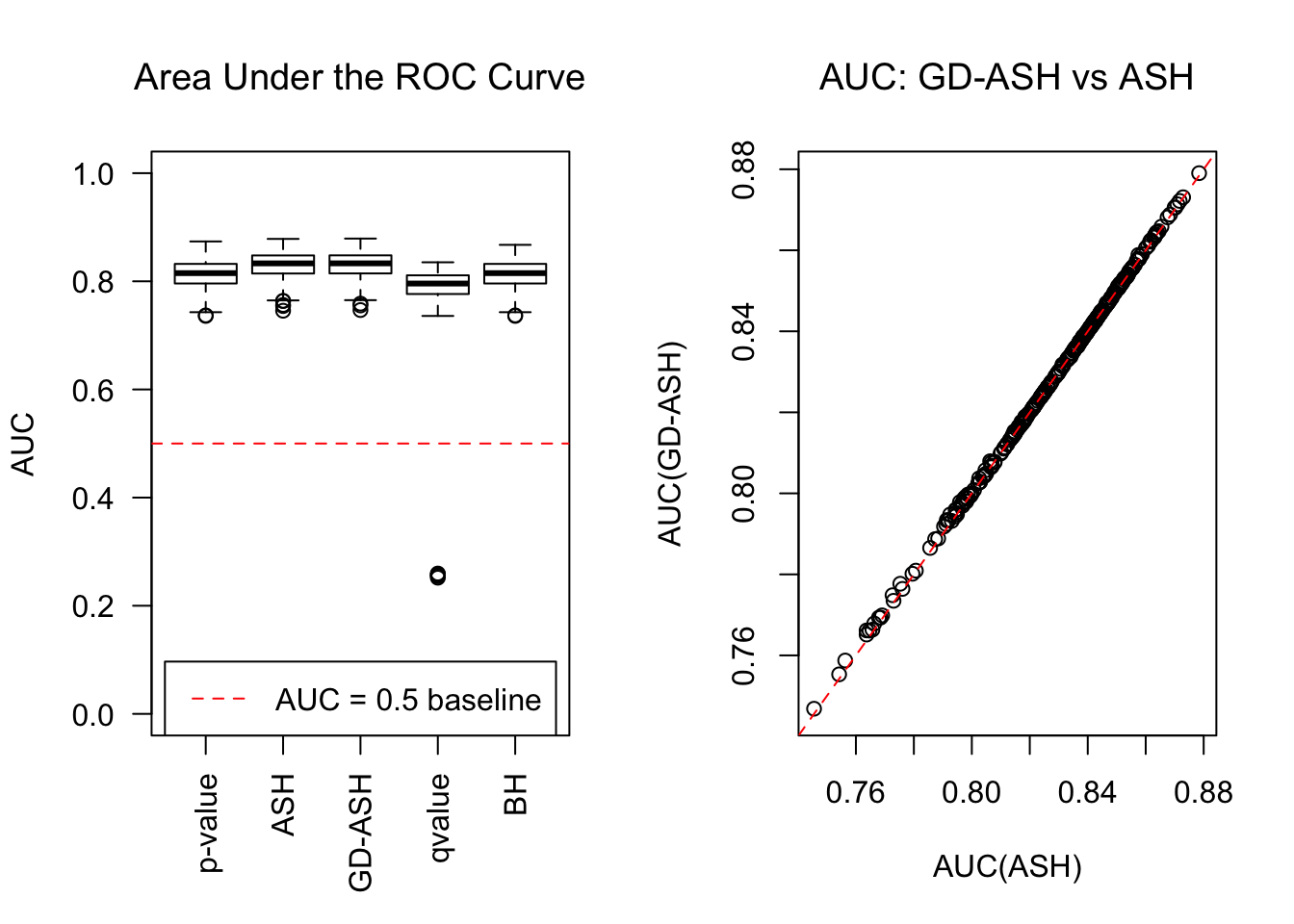
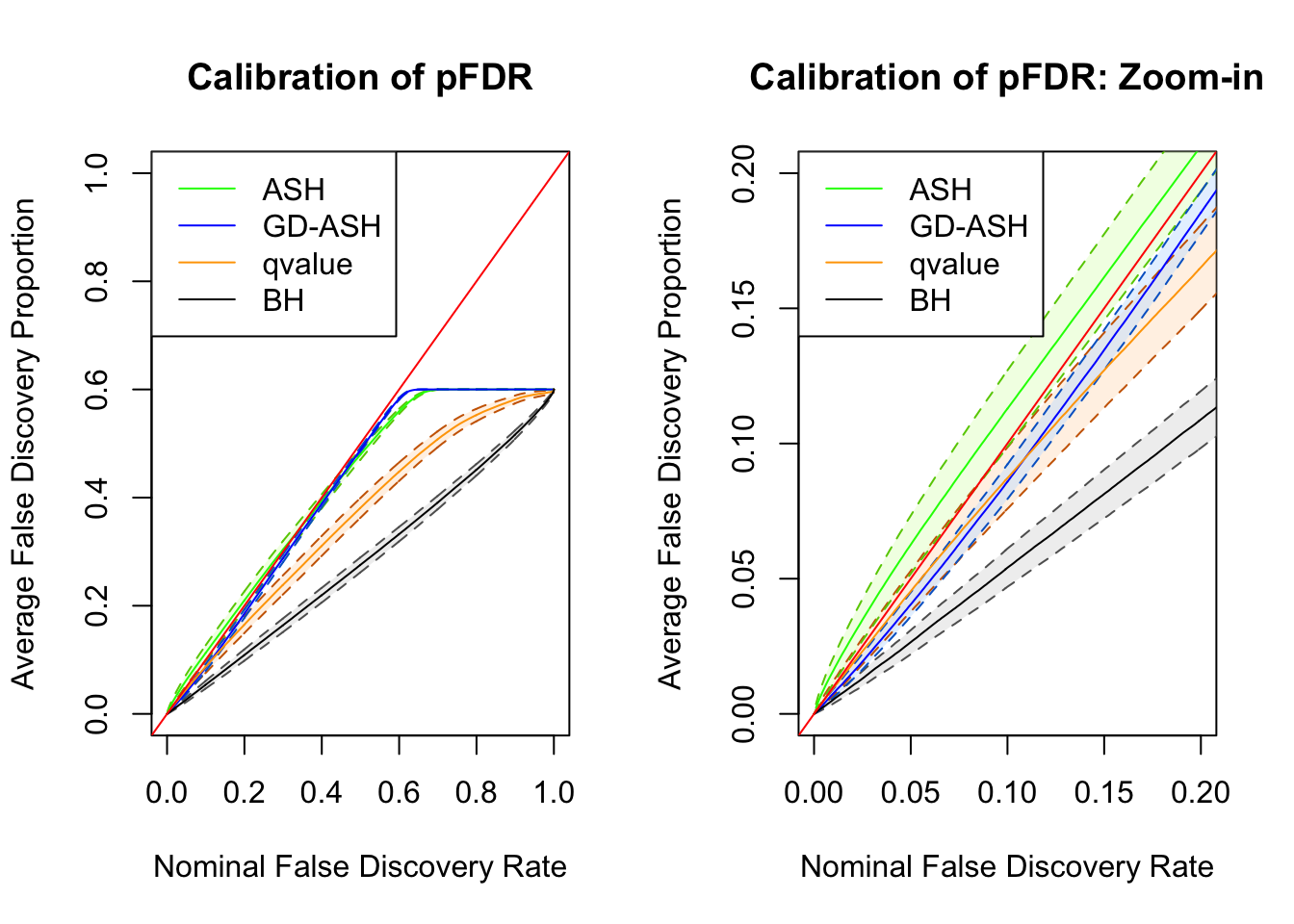
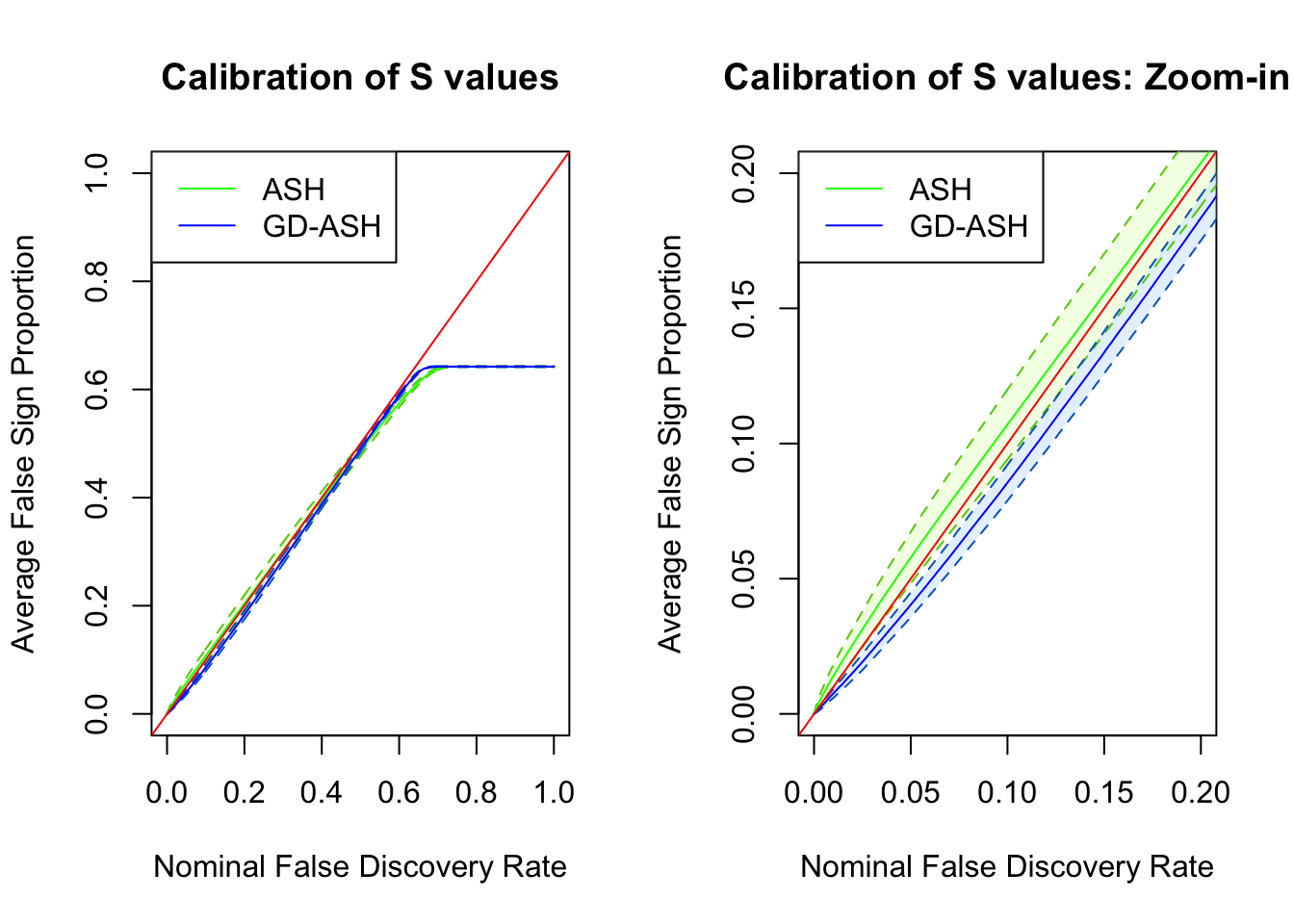
SNR = 10
\[ g = 0.6\delta_0 + 0.3N\left(0, \left(3\sigma\right)^2\right) + 0.1N\left(0, \left(8.54\sigma\right)^2\right) \ . \]
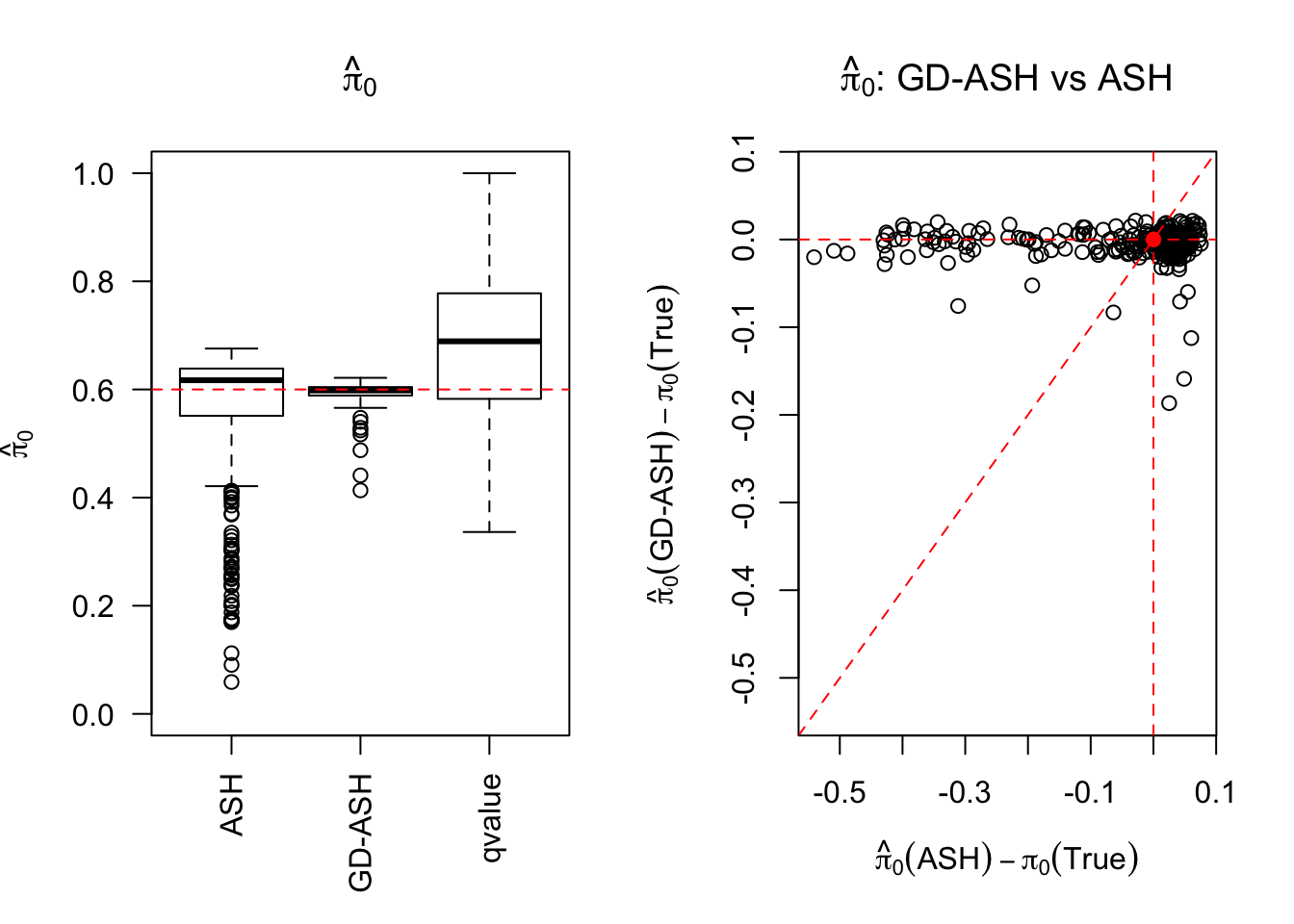
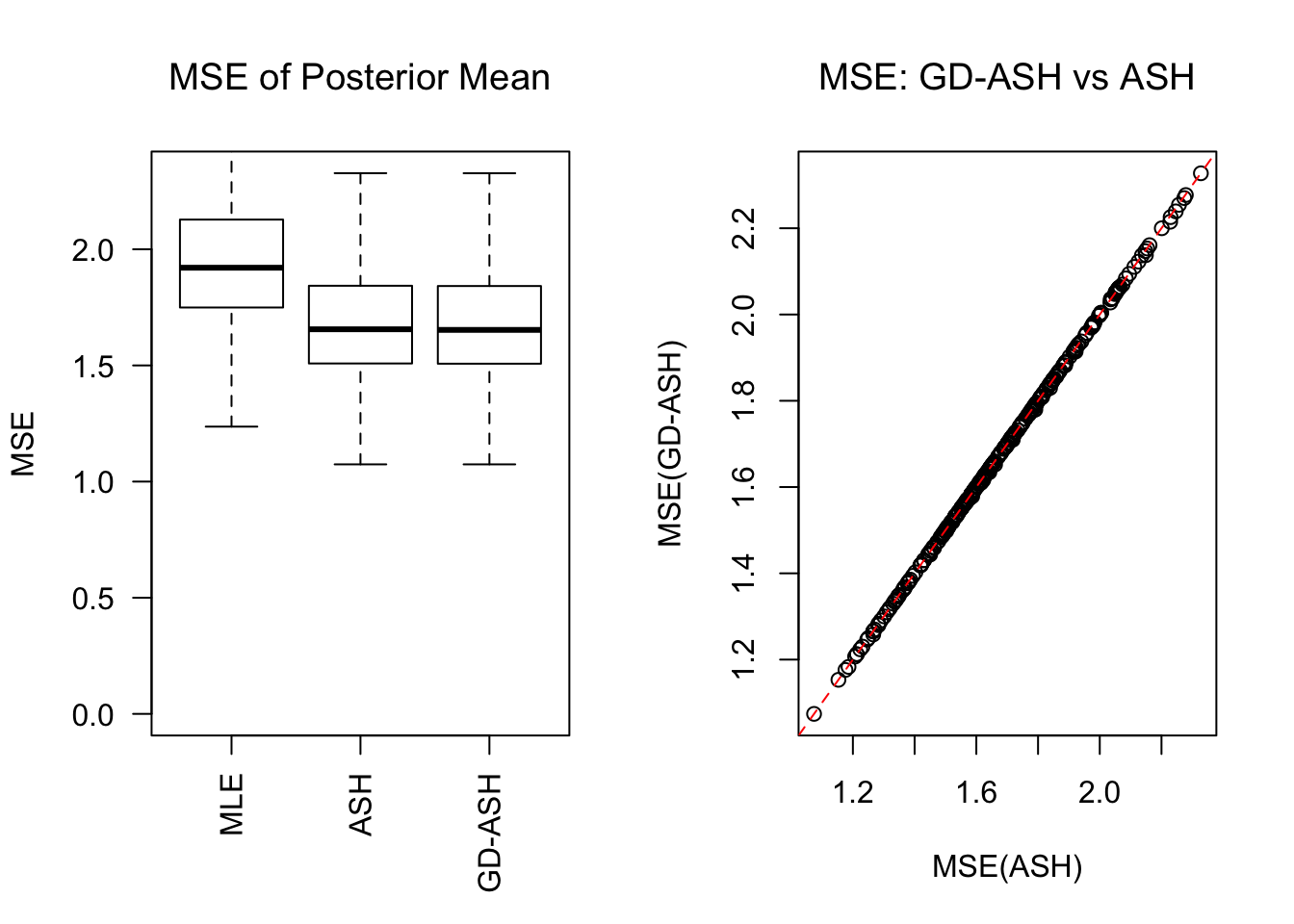
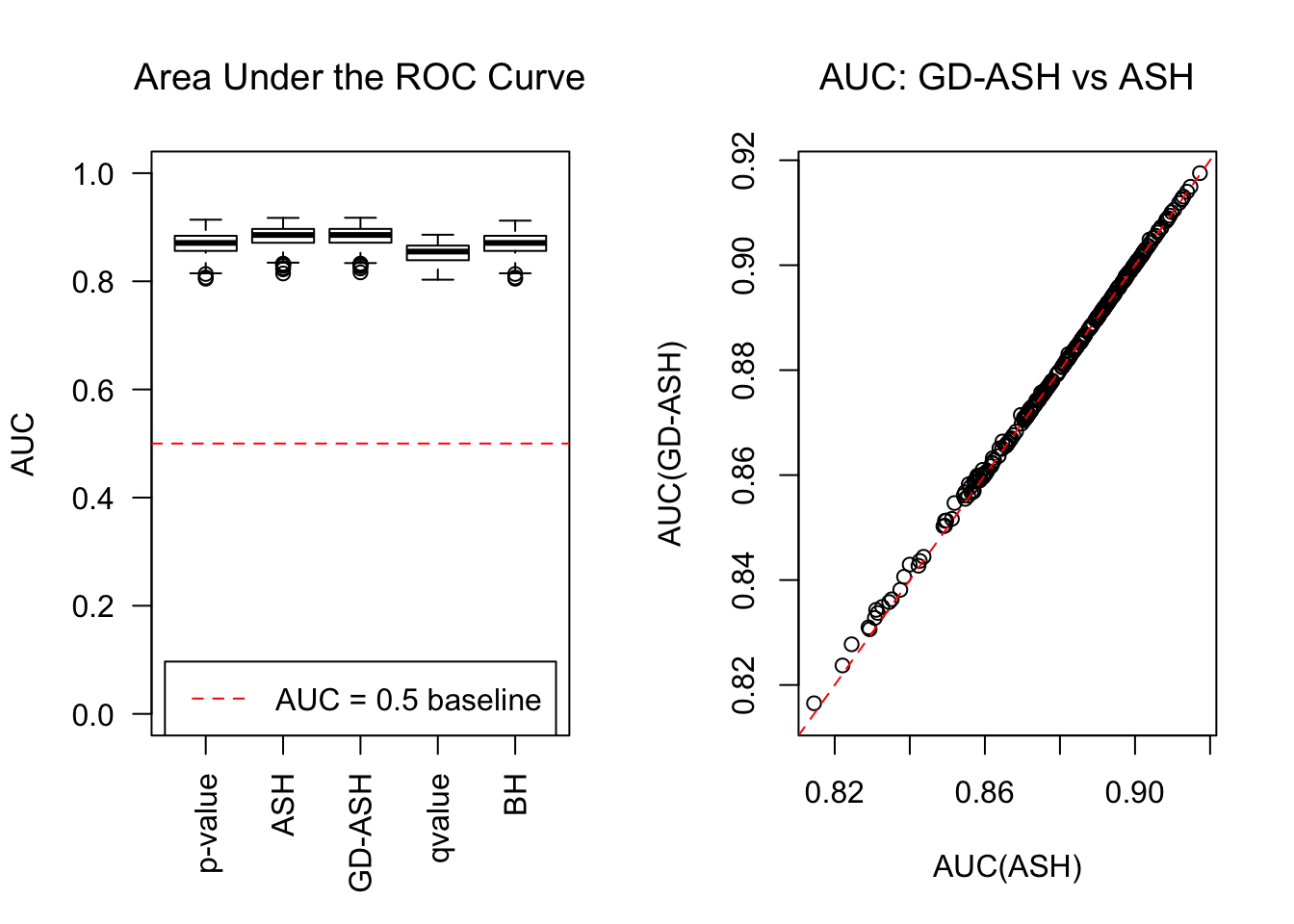
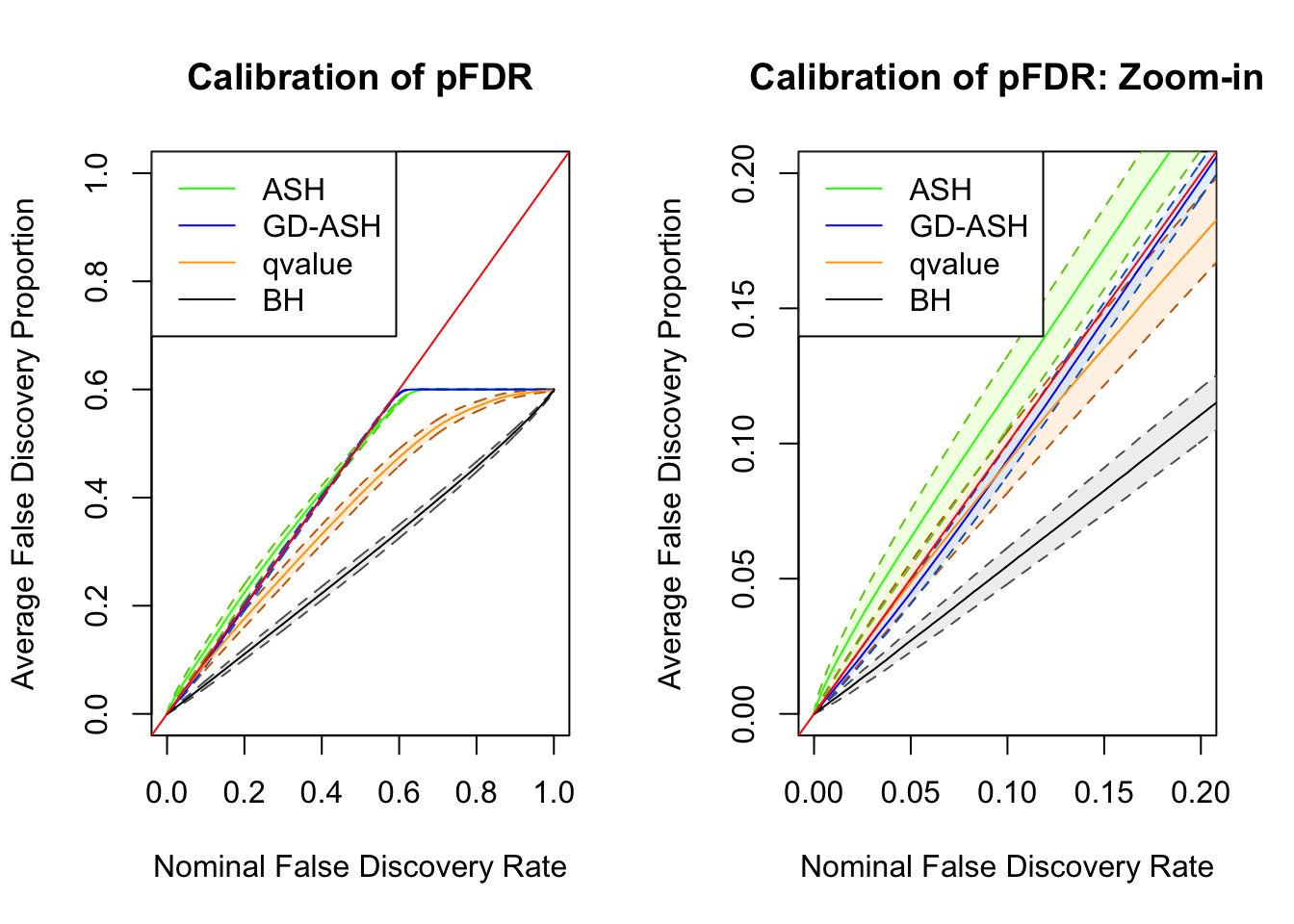
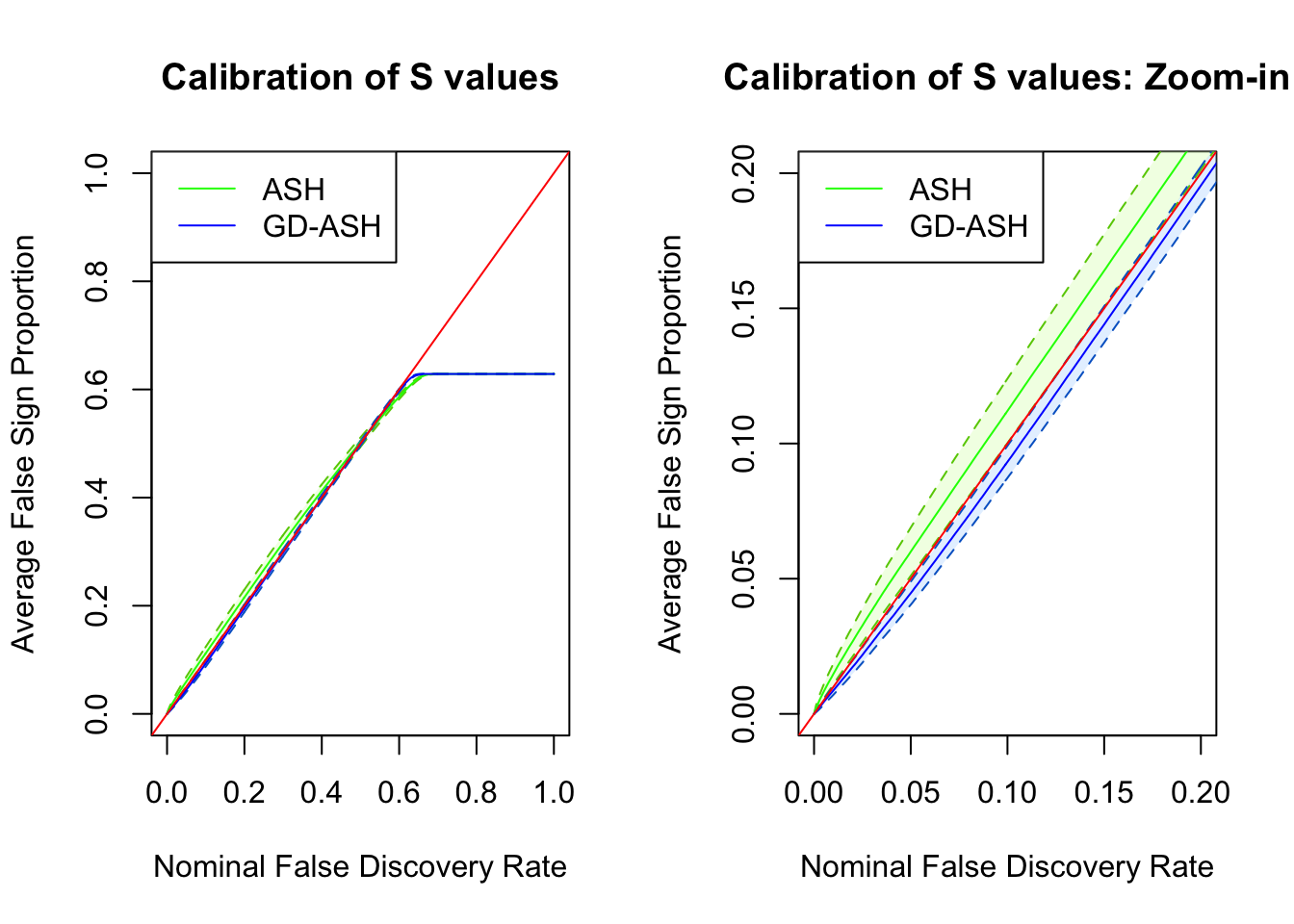
Session information
sessionInfo()R version 3.3.3 (2017-03-06)
Platform: x86_64-apple-darwin13.4.0 (64-bit)
Running under: macOS Sierra 10.12.5
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] Rmosek_7.1.2 PolynomF_0.94 cvxr_0.0.0.9009
[4] REBayes_0.85 Matrix_1.2-10 SQUAREM_2016.10-1
[7] EQL_1.0-0 ttutils_1.0-1 pROC_1.9.1
[10] edgeR_3.14.0 limma_3.28.21 ashr_2.1-13
loaded via a namespace (and not attached):
[1] Rcpp_0.12.10 knitr_1.16 magrittr_1.5
[4] MASS_7.3-47 doParallel_1.0.10 pscl_1.4.9
[7] lattice_0.20-35 foreach_1.4.3 plyr_1.8.4
[10] stringr_1.2.0 tools_3.3.3 parallel_3.3.3
[13] grid_3.3.3 git2r_0.18.0 htmltools_0.3.6
[16] iterators_1.0.8 yaml_2.1.14 rprojroot_1.2
[19] digest_0.6.12 codetools_0.2-15 evaluate_0.10
[22] rmarkdown_1.5 stringi_1.1.5 backports_1.0.5
[25] truncnorm_1.0-7 This R Markdown site was created with workflowr