To C or not to C, let Nim answer the question!
Table of content
- Introduction
- Looking for trouble
- Getting out of trouble
- Did I get out of trouble?
- Hello, is it Nim you're looking for?
Me, myself and I

Bonta Sergiu Vlad
Email/contact: bonta.vlad@gmail.com or just google my name
Web developer for about 3 years(it's less then 3 but not by much)
Me knows mostly Python and Javascript
“Do I really look like a guy with a plan? You know what I am? I'm a dog chasing cars. I wouldn't know what to do with one if I caught it! You know, I just… do things.”
― The Joker - Heath Ledger
Buyers beware
- I am you're average Joe programmer. No fancy big corporation, no big project, not much experience.
- Every benchmark out there lies, this included.
- I am very biased towards Nim.
“Madness, as you know, is a lot like gravity, all it takes is a little push” - The Joker
Looking for trouble
"Every Fairy Tale Needs A Good Old Fashioned Villain"
- cpu intensive
- relative easy to explain
- somehow similar to a real use case
The villain
We have around 1000 Truevision images that we want to open, fill the first 5 pixels with red color(simulate a watermark?) and finally save it.
Image used

Need for speed: Hot pursuit two!
It's a batch process
Each delay will add up!
Image sizes: 15, 150, 512, 1024, 2048 and 4096 pixels wide.
Truevision image format or *.tga
Often referred to as targa: raster graphics format.
Why tga?
- Simple compression algorithm.
- Used is computer graphics so it has some use cases.
- Found a python package that I could port(copy).
TGA structure
| header | pixel data | footer |
|---|---|---|
| id length | RLE | Extension offset |
| … | or | … |
| image specifications | no compression | Signature |
Pixel structure:
- Grayscale - 8 bit depth
- RGB - 16 bit depth
- RGB - 24 bit depth
- RGBA - 32 bit depth
RLE compression algorithm Run-length encoding (RLE)
lossless data compression
A hypothetical scan line, with B - black pixel and W - white:
WWWWBBBWWBBBBBBW
With a run-length encoding (RLE):
4W3B2W6B1W
Getting out of trouble
Now that we know our "villain" how can we overcome it?
“If I have to have a past, then I prefer it to be multiple choice.” ― Joker from Batman: The Killing Joke
Keep it simple stupid!
Maybe we can "cheat"!
We do not strive for the fastest implementation, we strive for "good enough" with minimal, preferably none, development time
cPython
The hero we need, the hero we want(well most of the time at least)!
The slowest implementation, but good reference point
Serves as blueprint for the Nim implementation
class TGAHeader(object): def __init__(self): self.id_length = 0 self.color_map_type = 0 self.image_type = 0 # ... class TGAFooter(object): def __init__(self): self.extension_area_offset = 0 self.developer_directory_offset = 0 self.signature = "TRUEVISION-XFILE" # ...
class Image(object): def __init__(self): self._header = TGAHeader() self._footer = TGAFooter() self._pixels = [] def load(self, file_name): pass # ... def save(self, file_name, original_format=False, force_16_bit=False, compress=False): pass # ... @staticmethod def _encode(row): """Econde a row of pixels. This function is a generator used during the compression phase. More information on packets generated are after returns section.""" pass # ...
Nuitka
Nuitka is a Python compiler. It's fully compatible with Python 2.7 .. 3.4.
You feed it your Python app, it does a lot of clever things, and spits out an executable or extension module.
If interpreting things is slow, why not compile it? Sounds crazy? think again!
nuitka --recurse-all program.py and you are set. recurse-all option will transverse the dependencies tree and compile them to, one by one.
Numba
With a few annotations, array-oriented and math-heavy Python code can be made to be similar in performance to C, C++ and Fortran.
Numba works by generating optimized machine code using the LLVM compiler.
Compilation can run on either CPU or GPU hardware, integrates well with Python scientific software stack.
# Taken directly from the project home page from numba import jit from numpy import arange # jit decorator tells Numba to compile this function. # The argument types will be inferred by Numba when function is called. @jit def sum2d(arr): M, N = arr.shape result = 0.0 for i in range(M): for j in range(N): result += arr[i,j] return result a = arange(9).reshape(3,3) print(sum2d(a))
Not included in the benchmark because:
The following Python language features are not currently supported:
- Function definition
- Class definition
- Exception handling (
try .. except,try .. finally) - Context management (the
withstatement) - Comprehensions (either
list,dict,setorgenerator comprehensions) - Generator delegation (
yield from)
PyPy
PyPy is a fast, compliant alternative implementation of the Python language (2.7.12 and 3.3.5). It has several advantages and distinct features, speed, memory usage, compatibility, stackless
Get a huge speed improvement by just replacing python with pypy
eg: pypy program.py. To good to be true? Yes, yes it is!
Two things: warmup time and incompatibility with all those good python modules written with the help of C.
Nim
Nim (formerly known as "Nimrod") is a statically typed, imperative programming language that tries to give the programmer ultimate power without compromises on runtime efficiency. This means it focuses on compile-time mechanisms in all their various forms.
Nim-pymod
- Auto-generates a Python module that wraps a Nim module
- pymod consists of Nim bindings & Python scripts to automate the generation of Python C-API extensions
- There's even a PyArrayObject that provides a Nim interface to Numpy arrays.
## Compile this Nim module using the following command: ## python path/to/pmgen.py greeting.nim ## Taken directly from the projects README import strutils # `%` operator import pymod import pymodpkg/docstrings proc greet*(audience: string): string {.exportpy.} = docstring"""Greet the specified audience with a familiar greeting. The string returned will be a greeting directed specifically at that audience. """ return "Hello, $1!" % audience initPyModule("hw", greet)
>>> import hw
>>> hw.greet
<built-in function greet>
>>> hw.greet("World")
'Hello, World!'
>>> help(hw.greet)
Help on built-in function greet in module hw:
greet(...)
greet(audience: str) -> (str)
Parameters
----------
audience : str -> string
Returns
-------
out : (str) <- (string)
Greet the specified audience with a familiar greeting.
The string returned will be a greeting directed specifically at that audience.
>>>
Procedure parameter & return types
The following Nim types are currently supported by Pymod:
| Type family | Nim types | Python2 type | Python3 type |
|---|---|---|---|
| floating-point | `float`, `float32`, `float64`, `cfloat`, `cdouble` | `float` | `float` |
| signed integer | `int`, `int16`, `int32`, `int64`, `cshort`, `cint`, `clong` | `int` | `int` |
| unsigned integer | `uint`, `uint8`, `uint16`, `uint32`, `uint64`, `cushort`, `cuint`, `culong`, `byte` | `int` | `int` |
| non-unicode character | `char`, `cchar` | `str` | `bytes` |
| string | `string` | `str` | `str` |
| Numpy array | `ptr PyArrayObject` | `numpy.ndarray` | `numpy.ndarray` |
Support for the following Nim types is in development:
| Type family | Nim types | Python2 type | Python3 type |
|---|---|---|---|
| signed integer | `int8` | `int` | `int` |
| boolean | `bool` | `bool` | `bool` |
| unicode code point (character) | `unicode.Rune` | `unicode` | `str` |
| non-unicode character sequence | `seq[char]` | `str` | `bytes` |
| unicode code point sequence | `seq[unicode.Rune]` | `unicode` | `str` |
| sequence of a single type T | `seq[T]` | `list` | `list` |
Going Commando: ditching pymod and using ctypes
Original blog post here: http://akehrer.github.io/posts/connecting-nim-to-python/
# median_test.nim proc median*(x: openArray[float]): float {. exportc, dynlib .} = ## Computes the median of the elements in `x`. ## If `x` is empty, NaN is returned. if x.len == 0: return NAN var sx = @x # convert to a sequence since sort() won't take an openArray sx.sort(system.cmp[float]) if sx.len mod 2 == 0: var n1 = sx[(sx.len - 1) div 2] var n2 = sx[sx.len div 2] result = (n1 + n2) / 2.0 else: result = sx[(sx.len - 1) div 2]
Python Code
from ctypes import * def main(): test_lib = CDLL('median_test') # Function parameter types test_lib.median.argtypes = [POINTER(c_double), c_int] # Function return types test_lib.median.restype = c_double # Calc some numbers nums = [1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0] nums_arr = (c_double * len(nums))() for i,v in enumerate(nums): nums_arr[i] = c_double(v) med_res = test_lib.median(nums_arr, c_int(len(nums_arr))) print('The median of %s is: %f'%(nums, med_res)) if __name__ == '__main__': main()
Compile and run
$nim c -d:release --app:lib median_test.nim $python median.py The median of [1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0] is: 4.500000
How do we know the arguments type?
nim c --app:lib --header median_test.nim
The --header option will produce a C header file in the nimcache folder where the module is compiled.
#ifndef __median_test__ #define __median_test__ #define NIM_INTBITS 32 #include "nimbase.h" N_NOCONV(void, signalHandler)(int sig); N_NIMCALL(NI, getRefcount)(void* p); N_LIB_IMPORT N_CDECL(NF, median)(NF* x, NI xLen0); N_LIB_IMPORT N_CDECL(void, NimMain)(void); #endif /* __median_test__ */
PyMod -> NimTga implementation
# ... proc loads*(filename: string): tuple[header, footer: string, pixels: ptr PyArrayObject] {.exportpy, returnDict.}= let image = newImage(filename) let shape = getShape(image.header) # pixel shape: 1, 3 or 4 values result.header = $$image.header result.footer = $$image.footer # create a numpy array [[...shape], [...shape]] result.pixels = createSimpleNew([image.pixels.high, shape], np_uint8) # fill them with zeroes to get rid of random values doFILLWBYTE(result.pixels, 0) var i = 0 for mval in result.pixels.accessFlat(uint8).mitems: # Forward-iterate through the array. mval = image.pixels[i div shape][i mod shape] inc(i)
proc saves*(header, footer: string, pixels: ptr PyArrayObject, filename: string, compress: int) {.discardable, exportpy.}= var image = newImage(to[Header](header), to[Footer](footer)) i = 0 let shape = pixels.strides[0] var pixel_data = newSeq[uint](shape) for v in pixels.accessFlat(uint8).items: pixel_data[i] = v if i == shape - 1: image.pixels.add(newPixel(pixel_data)) pixel_data = newSeq[uint](shape) i = -1 inc(i) image.save(filename, compress.bool) initPyModule("_ntga", loads, saves)
# ... class Image(object): def __init__(self, filename): _data = _ntga.loads(filename) self._header = json.loads(_data["header"]) self._footer = json.loads(_data["footer"]) for section in [self._header, self._footer]: for k, v in section.items(): setattr(self, k, v) self.pixels = _data["pixels"] def save(self, filename, compress=False): for section in [self._header, self._footer]: for k in section: section[k] = getattr(self, k) _ntga.saves(json.dumps(self._header), json.dumps(self._footer), self.pixels, filename, int(compress))
@property def pixel_size(self): if self._header['image_type'] in [3, 11]: return 1 elif self._header['image_type'] in [2, 10]: if self._header['pixel_depth'] == 16 or self._header['pixel_depth'] == 24: return 3 elif self._header['pixel_depth'] == 32: return 4 else: raise ValueError("Invalid pixel depth") else: raise ValueError("Invalid pixel depth")
def main(image_path=None): image_path = image_path if image_path else sys.argv[1] image = Image(image_path) for i in range(5): image.pixels[i] = pixel image.save("dump.tga", compress=True) if __name__ == "__main__": main()
Did I get out of trouble?
“Enough madness? Enough? And how do you measure madness? - The Joker”
PC specs
- Motherboard: Abit IP35
- CPU: Intel(R) Xeon(R) X5460 @ 3.16GHz 4 cores
- Memory: DDR2 4GiB @ 800MHz
- HDD: Seagate Baracuda x 2 RAID0
import os from subprocess import call BASE_PATH = os.path.join(os.getcwd(), 'images') def st_time(func): from functools import wraps import time @wraps(func) def st_func(*args, **kwargs): t1 = time.time() func(*args, **kwargs) t2 = time.time() return t2 - t1 return st_func @st_time def cpython(image_path): from pyTGA.measure import main return main(image_path) @st_time def pypy(image_path): return call(["pypy", "pyTGA/measure.py", image_path]) # ...
# start the x axis at 0 x = [0, 15, 150, 512, 1024, 2048, 4096] tests = [cpython, pypy, nuitka, nim, pymod_nim] images = ["pie_15_11.tga", "pie_150_113.tga", "pie_512_384.tga", "pie_1024_768.tga", "pie_2048_1536.tga", "pie_4096_3072.tga"] for t in tests: res = [0, ] # 0 because we want to start from the same point on both axis for image in images: image_path = os.path.join(BASE_PATH, image) # res will be ploted with matplotlib res.append(t(image_path)) print "benchmarking: {} with size: {}".format(t.__name__, image)
Results
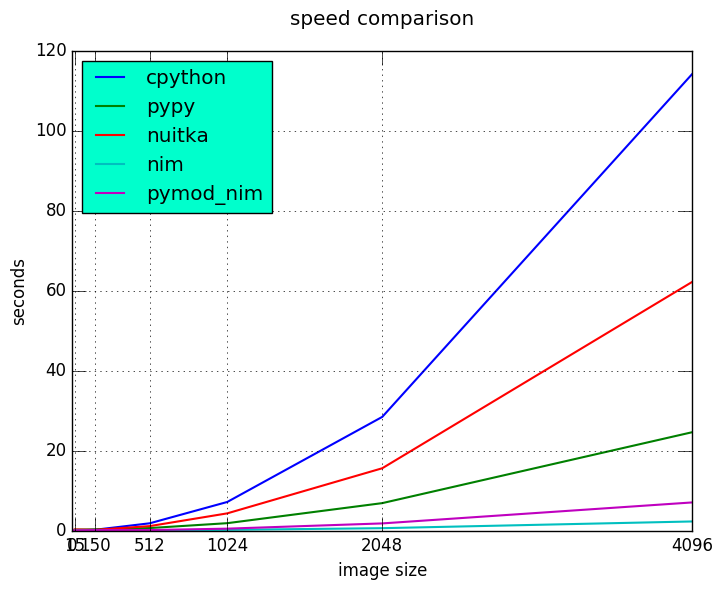
Detail view on the fastest three
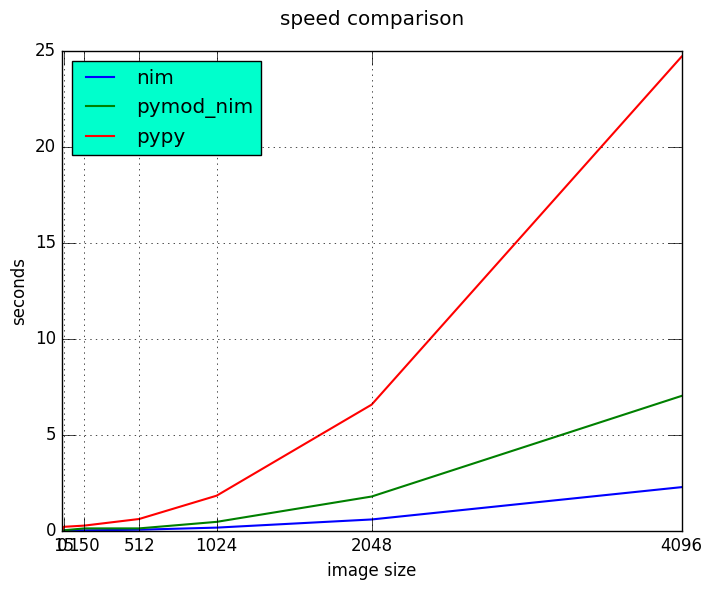
Detail view on small execution time
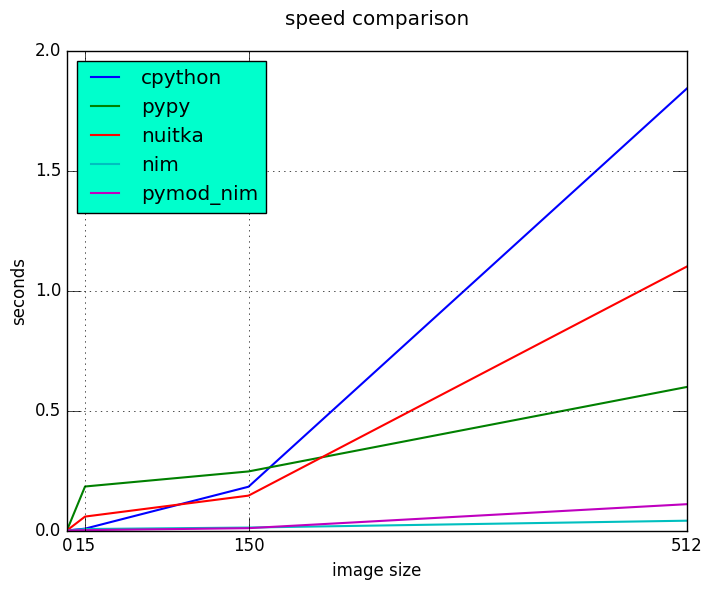
Hello, is it Nim you're looking for?
“True love is finding someone whose demons play well with yours” ― The Joker Batman Arkham City
What is Nim?
Original talk given by the language creator Andreas Rumpf at OSCON 2015
- new system programming language
- compiles to C
- garbage collection + manual memory management
- thread local garbage collection
- design goals: efficient, expressive, elegant
Goals
- as fast as C
- as expressive as Python
- as extensible as Lisp
Uses of Nim
- web development
- games
- compilers
- operating system development
- scientific computing
- scripting
- command line applications
- UI applications
- And lots more!
Nim for Python programmers
Similarities and differences.
| Feature | Python | Nim |
|---|---|---|
| Execution model | Virtual Machine, JIT | Machine code via C* |
| Meta-programming | Python (decorators/metaclasses/eval) | Nim (const/when/template/macro) |
| Memory Management | Garbage-collected | Garbage-collected and manual |
| Types | Dynamic | Static |
| Dependent types | - | Partial support |
| Generics | Duck typing | Yes |
| int8/16/32/64 types | No | Yes |
| Bigints (arbitrary size) | Yes (transparently) | Yes (via nimble package) |
| Arrays | Yes | Yes |
| Bounds-checking | Yes | Yes |
| Type inference | Duck typing | Yes (extensive support) |
| Closures | Yes | Yes |
| Operator Overloading | Yes | Yes (on any type) |
| Custom Operators | No | Yes |
| Object-Oriented | Yes | Minimalistic** |
| Methods | Yes | Yes |
| Multi-Methods | No | Yes |
| Exceptions | Yes | Yes |
\*Other backends supported and/or planned \** Can be achieved with macros
" The real joke is your stubborn, bone deep conviction that somehow, somewhere, all of this makes sense! That's what cracks me up each time!" - Batman #681
Bonta Sergiu Vlad
Email: bonta.vlad@gmail.com

Figure 6: https://bontavlad.github.io

Figure 7: https://github.com/BontaVlad/nimtga